Chapter 15. Meiosis & Sexual Reproduction

Chapter Outline
- 15.1 The Process of Meiosis
- 15.2 Disorders in Chromosome Number
- 15.3 Sexual Reproduction
Introduction
The ability to reproduce in kind is a basic characteristic of all living things. In kind means that the offspring of any organism closely resemble their parents. Hippopotamuses give birth to hippopotamus calves, Joshua trees produce seeds from which Joshua tree seedlings emerge, and adult flamingos lay eggs that hatch into flamingo chicks. In kind does not generally mean exactly the same. Whereas many unicellular organisms and a few multicellular organisms can produce genetically identical clones of themselves through cell division, many single-celled organisms and most multicellular organisms reproduce sexually. Sexual reproduction is the production by parents of two haploid cells and the fusion of two haploid cells to form a single, unique diploid cell. In most plants and animals, through tens of rounds of mitotic cell division, this diploid cell will develop into an adult organism. Haploid cells that are part of the sexual reproductive cycle are produced by a type of cell division called meiosis. The vast majority of eukaryotic organisms employ some form of meiosis and fertilization to reproduce. The evolutionary success of sexual reproduction may result from the genetic variation that it introduces into a species’ genome.
15.1 | The Process of Meiosis
Learning Objectives
By the end of this section, you will be able to:
- Describe the behavior of chromosomes during meiosis.
- Describe cellular events during meiosis.
- Explain the differences between meiosis and mitosis.
- Explain the mechanisms within meiosis that generate genetic variation among the products of meiosis.
Sexual reproduction requires fertilization, the union of two cells from two individual organisms. If those two cells each contain one set of chromosomes, then the resulting cell contains two sets of chromosomes. Cells containing one set of chromosomes are called haploid. Cells containing two sets of chromosomes are called diploid. If the reproductive cycle is to continue, then diploid cells must somehow reduce the number of chromosome sets before fertilization can occur again, or the number of chromosome will double in every generation. So, sexual reproduction requires a nuclear division that reduces the number of chromosome sets.
Most animals and plants are diploid. In each somatic (non-reproductive) cell of the organism, the nucleus contains two copies of each chromosome. The two copies of a chromosome are called homologous chromosomes, or homologs. Homologous means “similar”; homologous chromosomes are not identical, since they come from different parents. However, homologous chromosomes are matched pairs that contain the same genes in identical locations along their length. Diploid organisms inherit one copy of each homologous chromosome from each parent. For example, your body cells each contain one set of 23 chromosomes from your mother and one set of 23 chromosomes from your mother. Chromosome 1 from your mother and chromosome 1 from your father are homologous to each other.
Haploid cells are found only within structures that give rise to reproductive cells, such as gametes or spores. Spores are haploid cells that can produce a haploid organism or can fuse with another spore to form a diploid cell. All animals and most plants produce eggs and sperm, or gametes. Some plants and all fungi produce spores.
Meiosis is the nuclear division that forms haploid cells. Meiosis employs many of the same mechanisms as mitosis. However, whereas mitosis produces two genetically identical diploid daughter cells, meiosis produces four genetically different haploid daughter cells. To achieve this reduction in chromosome number, meiosis consists of two rounds of nuclear division. Because the events that occur during each of the division stages are analogous to the events of mitosis, the same stage names are assigned. However, because there are two rounds of division, the major process and the stages are designated with “I” or a “II.” Thus, meiosis I is the first round of meiotic division and consists of prophase I, prometaphase I, and so on. Meiosis II, in which the second round of meiotic division takes place, includes prophase II, prometaphase II, and so on.
15.1.1 Meiosis I
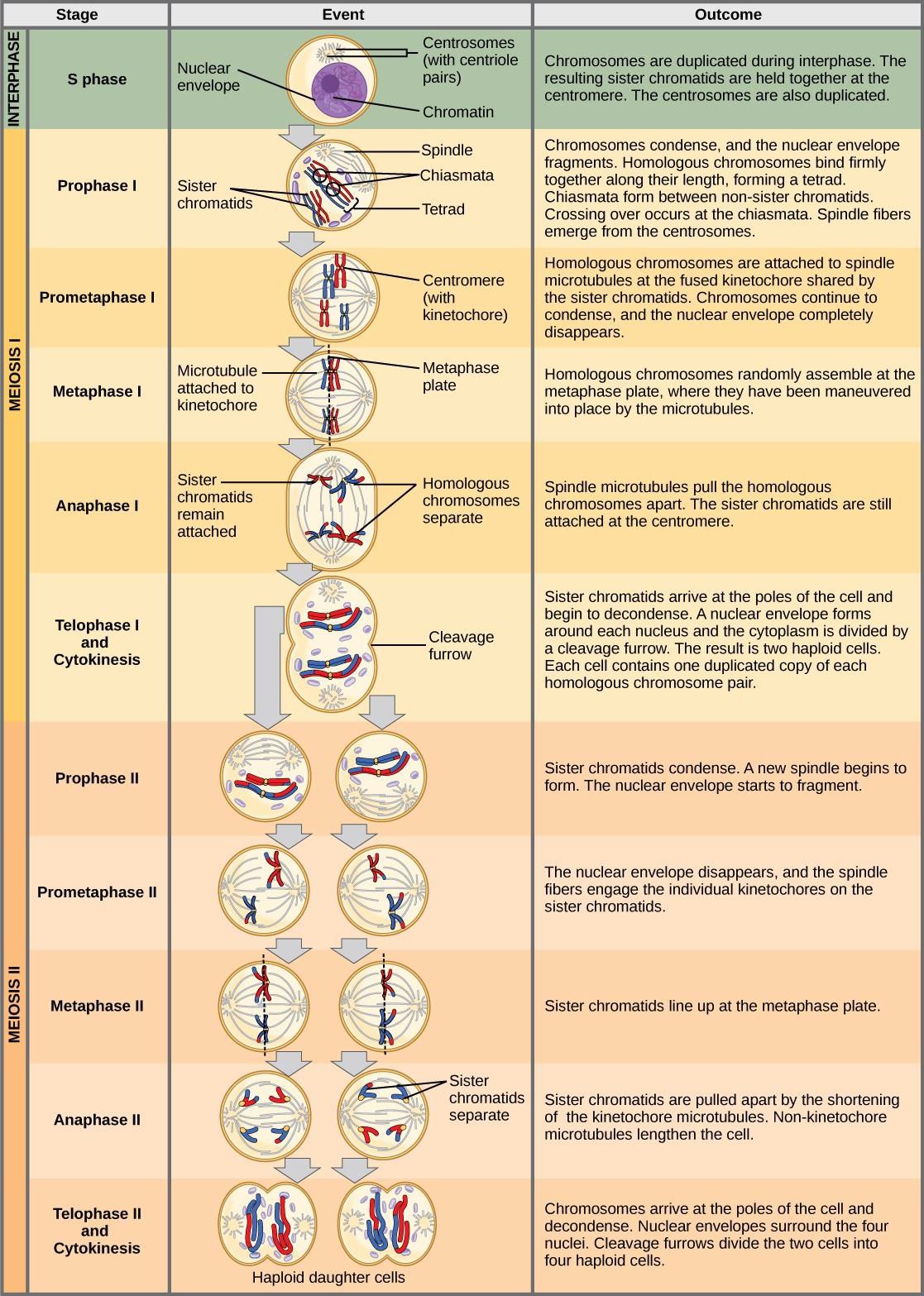
Meiosis is preceded by an interphase consisting of the G1, S, and G2 phases, which are nearly identical to the phases preceding mitosis. The G1 phase, which is also called the first gap phase, is the first phase of the interphase and is focused on cell growth. The S phase is the second phase of interphase, during which the DNA of the chromosomes is replicated. Finally, the G2 phase, also called the second gap phase, is the third and final phase of interphase; in this phase, the cell undergoes the final preparations for meiosis.
During DNA duplication in the S phase, each chromosome is replicated to produce two identical copies, called sister chromatids, that are held together at the centromere by cohesin proteins. Cohesin holds the chromatids together until anaphase II. The centrosomes, which are the structures that organize the microtubules of the meiotic spindle, also replicate. This prepares the cell to enter prophase I, the first meiotic phase.
Prophase I
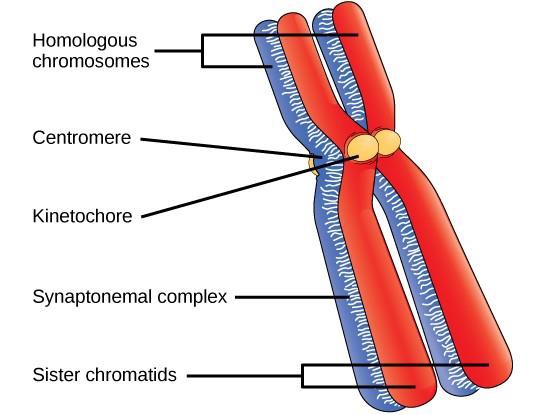
Early in prophase I, before the chromosomes can be seen clearly microscopically, the homologous chromosomes are attached at their tips to the nuclear envelope by proteins. As the nuclear envelope begins to break down, the proteins associated with homologous chromosomes bring the pair close to each other. Recall that in mitosis homologous chromosomes do not pair with each other. In mitosis, homologous chromosomes line up end-to-end, held together by a lattice of proteins called the synaptonemal complex. This tight pairing of the homologous chromosomes is called synapsis. In synapsis, the genes on the chromatids of the homologous chromosomes are aligned precisely with each other. (Figure 15.3).
In species such as humans, even though the X and Y sex chromosomes are not homologous (most of their genes differ), they have a small region of homology that allows the X and Y chromosomes to pair up during prophase I. A partial synaptonemal complex develops only between the regions of homology.
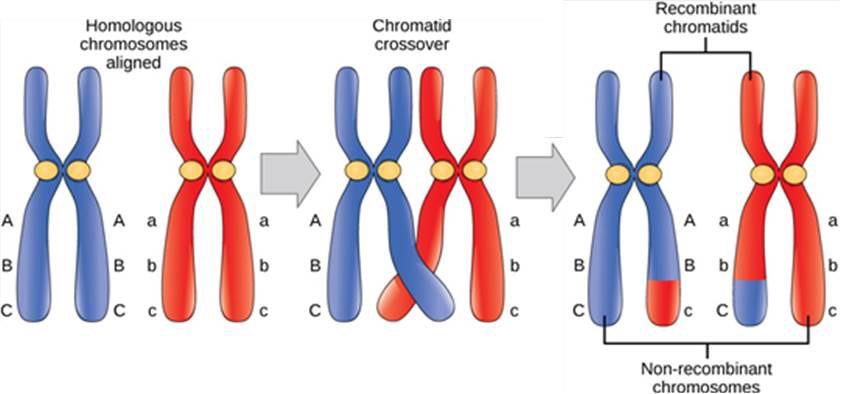
The synaptonemal complex allows the exchange of chromosomal segments between homologous chromatids, a process called crossing over or crossover. Crossing over can be observed visually under a microscope as chiasmata (singular = chiasma) (Figure 15.4). The number of chiasmata varies according to the species and the length of the chromosome. There must be at least one chiasma per chromosome for proper separation of homologous chromosomes during meiosis I, but there may be as many as 25.
As prophase I progresses, the synaptonemal complex begins to break down and the chromosomes begin to condense. At the end of prophase I, the pairs are held together only at the chiasmata and are called tetrads because the four sister chromatids of each pair of homologous chromosomes are now visible.
Crossover is the first source of genetic variation produced by meiosis. Crossover between a pair of homologous chromatids leads to a reciprocal exchange of equivalent DNA between a maternal chromosome and a paternal chromosome. When that sister chromatid is moved into a gamete cell, it will carry some DNA from one parent of the individual and some DNA from the other parent. This combination of maternal and paternal genes did not exist before the crossover.
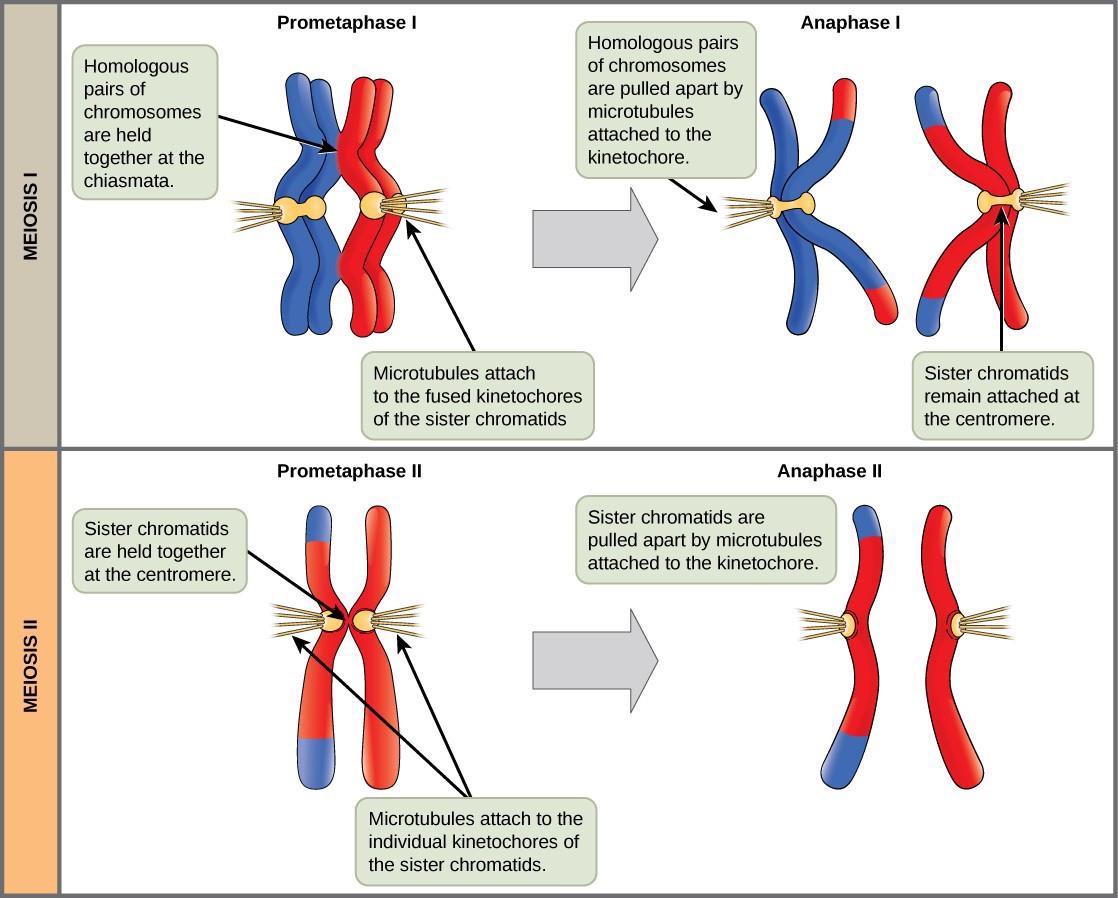
Prometaphase I
The key event in prometaphase I is the attachment of the spindle fiber microtubules to the kinetochore proteins at the centromeres. The microtubules move toward the middle of the cell and attach to one of the two fused homologous chromosomes. At the end of prometaphase I, each tetrad is attached to microtubules from both poles, with one homologous chromosome facing each pole. The homologous chromosomes are still held together at chiasmata. In addition, the nuclear membrane has broken down entirely.
Metaphase I
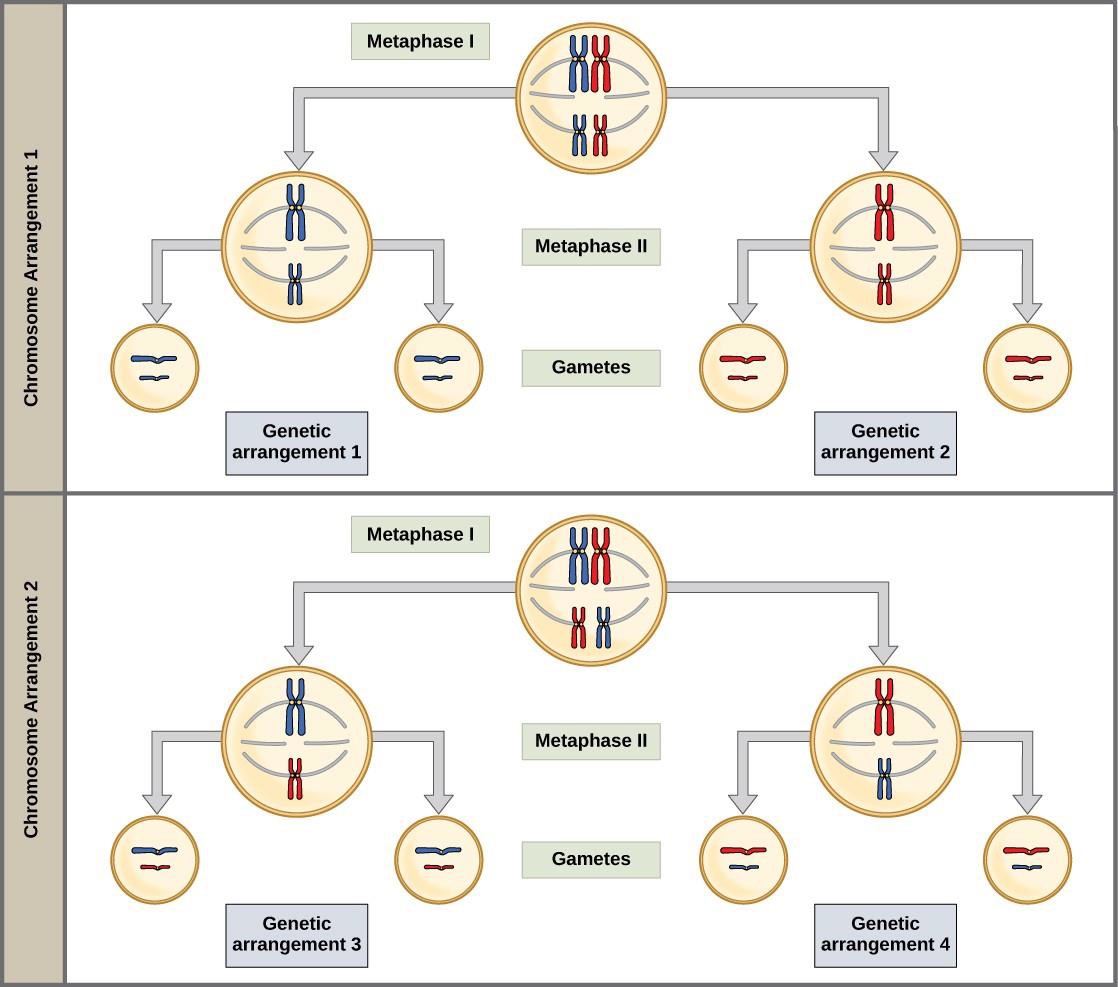
During metaphase I, the homologous chromosomes are arranged in the center of the cell with the kinetochores facing opposite poles. The homologous pairs orient themselves randomly at the equator. Because there is an equal chance that a microtubule fiber will encounter a maternally or paternally inherited chromosome, the arrangement of the tetrads at the metaphase plate is random. Any maternally inherited chromosome may face either pole. Any paternally inherited chromosome may also face either pole. The orientation of each tetrad is independent of the orientation of the other tetrads. This is important in determining the genes carried by a gamete, as each will only receive one of the two homologous chromosomes.
The random orientation of homologous chromosomes creates the second form of genetic variation in offspring. Recall that homologous chromosomes contain slight differences in their genetic information. In each cell that undergoes meiosis, the arrangement of the tetrads is different. Since humans have 23 chromosome pairs, there are over eight million possible genetically-distinct gametes. This number does not include the variability that was previously created in the sister chromatids by crossover. Given these two mechanisms, it is highly unlikely that any two haploid cells resulting from meiosis will have the same genetic composition (Figure 15.5).
Anaphase I
In anaphase I, the microtubules pull the linked chromosomes apart. The sister chromatids remain tightly bound together at the centromere. The chiasmata are broken in anaphase I as the microtubules attached to the fused kinetochores pull the homologous chromosomes apart (Figure 15.6).
Telophase I and Cytokinesis
In telophase, the separated chromosomes arrive at opposite poles. The remainder of the typical telophase events may or may not occur, depending on the species. In some organisms, the chromosomes decondense and nuclear envelopes form around the chromatids in telophase I. In other organisms, cytokinesis—the physical separation of the cytoplasmic components into two daughter cells—occurs without reformation of the nuclei. In nearly all species of animals and some fungi, cytokinesis separates the cell contents via a cleavage furrow (constriction of the actin ring that leads to cytoplasmic division). In plants, a cell plate is formed during cell cytokinesis by Golgi vesicles fusing at the metaphase plate. This cell plate will ultimately lead to the formation of cell walls that separate the two daughter cells.
Two haploid cells are the end result of the first meiotic division. The cells are haploid because at each pole, there is just one of each pair of the homologous chromosomes. However, each homolog still consists of two sister chromatids. Recall that sister chromatids are merely duplicates of one of the two homologous chromosomes (except for changes that occurred during crossing over). In meiosis II, these two sister chromatids will separate, creating four haploid daughter cells.
15.1.2 Meiosis II
In some species, cells enter a brief interphase, or interkinesis, before entering meiosis II. Interkinesis lacks an S phase, so chromosomes are not duplicated. The two cells produced in meiosis I go through the events of meiosis II in synchrony. During meiosis II, the sister chromatids within the two daughter cells separate, forming four new haploid gametes. The mechanics of meiosis II is similar to mitosis, except that each dividing cell has only one set of homologous chromosomes. Therefore, each cell has half the number of sister chromatids to separate out as a diploid cell undergoing mitosis.
Prophase and Prometaphase II
If the chromosomes decondensed in telophase I, they condense again. If nuclear envelopes were formed, they fragment into vesicles. The centrosomes that were duplicated during interkinesis move away from each other toward opposite poles, and new spindles are formed. The nuclear envelopes are completely broken down, and the spindle is fully formed. Each sister chromatid forms an individual kinetochore that attaches to microtubules from opposite poles.
Metaphase II
The sister chromatids are maximally condensed and aligned at the equator of the cell.
Anaphase II
The sister chromatids are pulled apart by the kinetochore microtubules and move toward opposite poles. Non-kinetochore microtubules elongate the cell. Note that during meiosis I, the homologous chromosomes are pulled apart, while in meiosis II, sister chromatids are pulled apart (Figure 15.6).
Telophase II and Cytokinesis
The chromosomes arrive at opposite poles and begin to decondense. Nuclear envelopes form around the chromosomes. Cytokinesis separates the two cells into four unique haploid cells. At this point, the newly formed nuclei are both haploid. The cells produced are genetically unique because of the random assortment of paternal and maternal homologs and because of the recombining of maternal and paternal segments of chromosomes (with their sets of genes) that occurs during crossover. The entire process of meiosis is outlined in Figure 15.7.
15.1.3 Comparing Meiosis and Mitosis
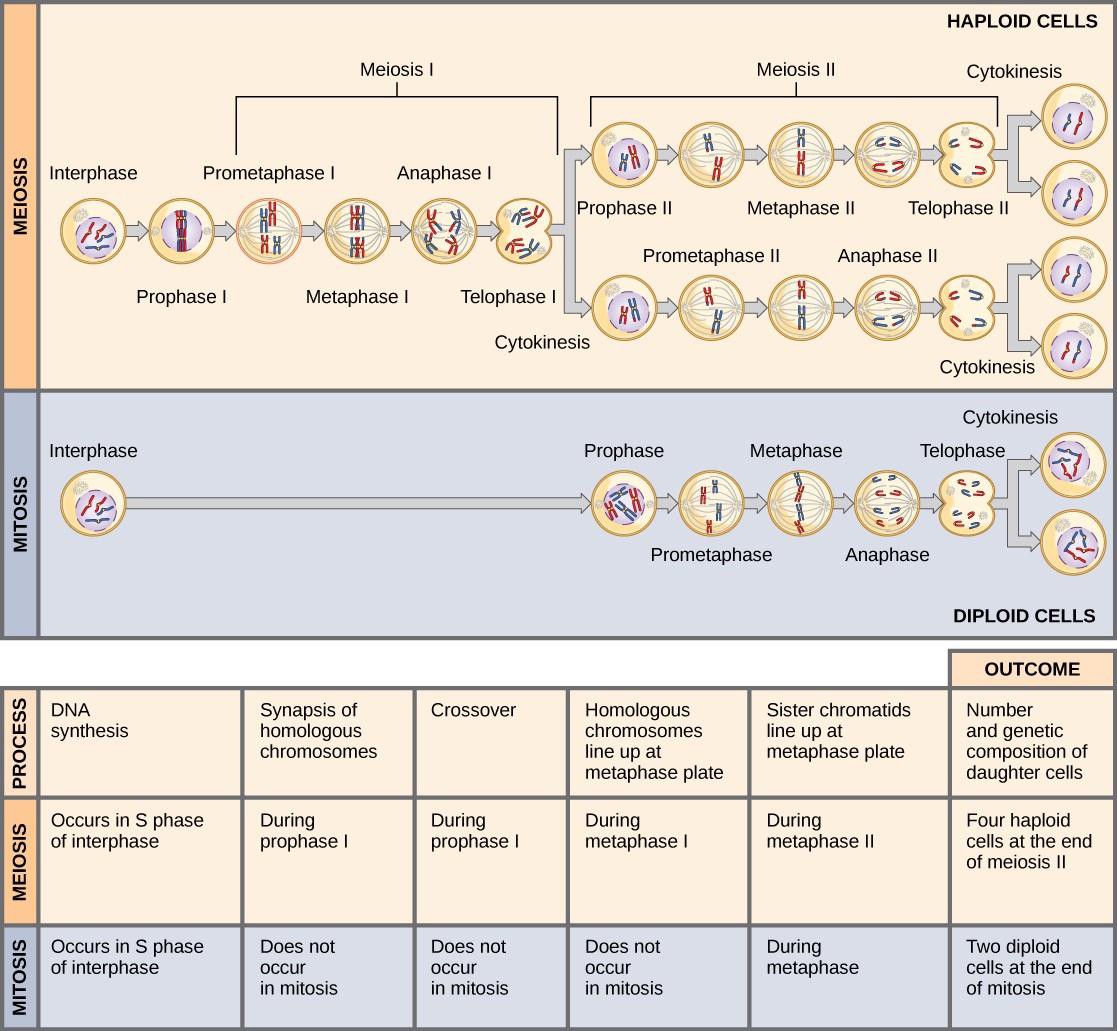
Mitosis and meiosis are both forms of division of the nucleus in eukaryotic cells. They share some similarities, but also exhibit distinct differences that lead to very different outcomes (Figure 15.8). Mitosis is a single nuclear division that results in two nuclei that are usually partitioned into two new cells. The nuclei resulting from a mitotic division are genetically identical to the original nucleus. They have the same number of sets of chromosomes. In most plants and all animal species, diploid cells typically undergo mitosis to form new diploid cells. In contrast, meiosis consists of two nuclear divisions resulting in four nuclei that are usually partitioned into four new cells. The nuclei resulting from meiosis are not genetically identical and they contain one chromosome set only. This is half the number of chromosome sets in the original diploid cell.
The main differences between mitosis and meiosis occur in meiosis I. In meiosis I, homologous chromosome pairs are bound together, undergo crossover, and line up randomly along the metaphase plate. When the homologs separate, the ploidy level is reduced from two to one. For this reason, meiosis I is referred to as a reduction division. There is no such reduction in ploidy level during mitosis.
Meiosis II is much more similar to mitosis. Duplicated chromosomes (only one set of them) line up on the metaphase plate and sister chromatids are separated to opposite poles. Meiosis II is not a reduction division because although there are fewer copies of the genome in the resulting cells, there is still one set of chromosomes, as there was at the end of meiosis I (Figure 15.8).
15.2 | Disorders in Chromosome Number
Learning Objectives
By the end of this section, you will be able to:
- Describe how a karyotype is made and used.
- Explain how nondisjunction occurs during meiosis.
- Describe common types of aneuploidy in humans.
- Explain why X chromosome nondisjunction causes less serious effects.
- Describe common chromosome structural rearrangements
15.2.1 Identification of Chromosomes
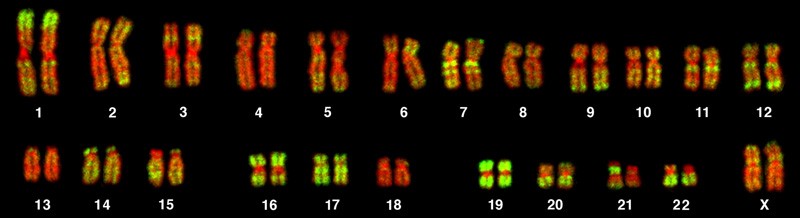
A karyotype is the number and appearance of chromosomes, and includes their length, banding pattern, and centromere position. To obtain a view of an individual’s karyotype, cytologists photograph the chromosomes and then cut and paste each chromosome into a chart, or karyogram (Figure 15.9).
Geneticists Use Karyograms to Identify Chromosomal Aberrations
Although Mendel is referred to as the “father of modern genetics,” he performed his experiments with none of the tools that the geneticists of today routinely employ. One such powerful cytological technique is karyotyping, a method in which traits characterized by chromosomal abnormalities can be identified from a single cell. To observe an individual’s karyotype, a person’s cells (such as white blood cells) are first collected from a blood sample or other tissue. In the laboratory, the isolated cells are stimulated to begin actively dividing. A chemical called colchicine is then applied to cells to arrest condensed chromosomes in metaphase. Cells are then made to swell using a hypotonic solution so the chromosomes spread apart. Finally, the sample is preserved in a fixative and applied to a slide.
The geneticist then stains chromosomes with one of several dyes to better visualize the distinct and reproducible banding patterns of each chromosome pair. Following staining, the chromosomes are viewed using bright-field microscopy. A common stain choice is the Giemsa stain. Giemsa staining results in approximately 400–800 bands (of tightly coiled DNA and condensed proteins) arranged along all of the 23 chromosome pairs; an experienced geneticist can identify each band. Chromosomes are further identified on the basis of size and centromere location. The geneticist obtains a digital image, identifies each chromosome, and manually arranges the chromosomes into a pattern (Figure 15.8).
At its most basic, the karyogram may reveal genetic abnormalities in which an individual has too many or too few chromosomes per cell. Examples of this are Down Syndrome, which is identified by a third copy of chromosome 21, and Turner Syndrome, which is characterized by the presence of only one X chromosome in women instead of the normal two. Geneticists can also identify large deletions or insertions of DNA. For instance, Jacobsen Syndrome—which involves distinctive facial features as well as heart and bleeding defects—is identified by a deletion on chromosome 11. Finally, the karyotype can pinpoint translocations, which occur when a segment of genetic material breaks from one chromosome and reattaches to another chromosome or to a different part of the same chromosome. Translocations are implicated in certain cancers, including chronic myelogenous leukemia.
During Mendel’s lifetime, inheritance was an abstract concept that could only be inferred by performing crosses and observing the traits expressed by offspring. By observing a karyogram, today’s geneticists can actually visualize the chromosomal composition of an individual to confirm or predict genetic abnormalities in offspring, even before birth.
In a given species, chromosomes can be identified by their number, size, centromere position, and banding pattern. In a human karyotype, autosomes (all of the non–sex chromosomes) are organized from largest (chromosome 1) to smallest (chromosome 22). The X and Y chromosomes are not autosomes. The chromosome “arms” projecting from either end of the centromere may be designated as short or long, depending on their relative lengths. The short arm is abbreviated p (for “petite”), whereas the long arm is abbreviated q (because it follows “p” alphabetically). Each arm is further subdivided and denoted by a number. Using this naming system, locations on chromosomes can be described consistently in the scientific literature.
15.2.2 Nondisjunction during Meiosis Leads to Aneuploidy
Of all of the chromosomal disorders, abnormalities in chromosome number are the most obviously identifiable. Disorders of chromosome number include the duplication or loss of entire chromosomes, as well as changes in the number of complete sets of chromosomes. They are caused by nondisjunction, which occurs when pairs of homologous chromosomes or sister chromatids fail to separate during meiosis. Misaligned or incomplete synapsis, or a dysfunction of the spindle apparatus that facilitates chromosome migration, can cause nondisjunction. The risk of nondisjunction increases with the age of the parents.
Nondisjunction can occur during either meiosis I or II (Figure 15.10). If homologous chromosomes fail to separate during meiosis I, the result is two gametes that lack that particular chromosome and two gametes with two copies of the chromosome. If sister chromatids fail to separate during meiosis II, the result is one gamete that lacks that chromosome, two normal gametes with one copy of the chromosome, and one gamete with two copies of the chromosome.
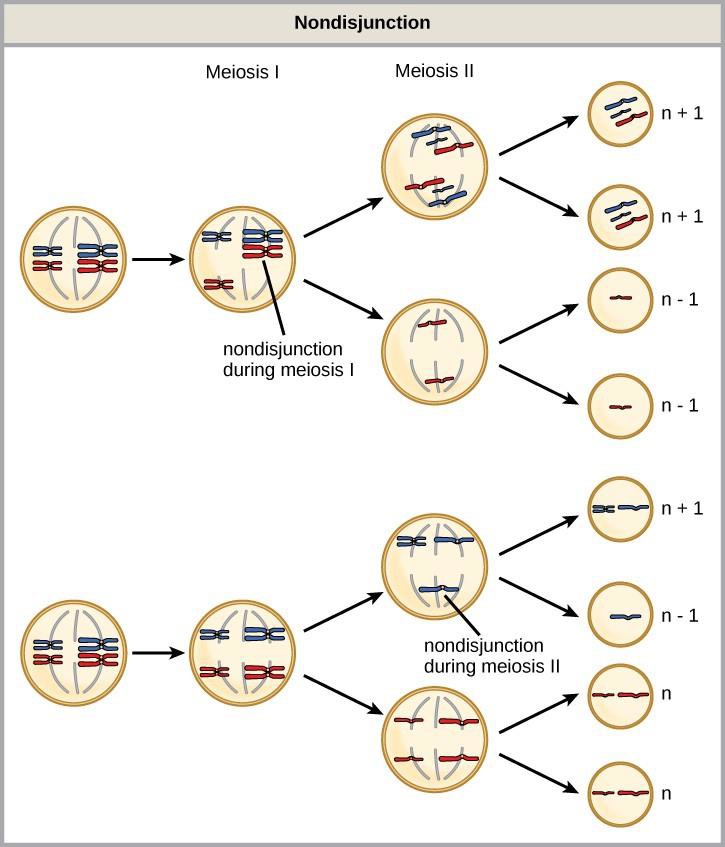
Aneuploidy
An individual with the appropriate number of chromosomes for their species is called euploid; in humans, euploidy corresponds to 22 pairs of autosomes and one pair of sex chromosomes. An individual with an error in chromosome number is described as aneuploid.
If a gamete that lacks a chromosome due to nondisjunction during meiosis is fertilized, the resulting individual will have only one copy of that chromosome, a condition referred to as monosomy. If a gamete with an extra copy of a chromosome is fertilized, the resulting individual will have three copies of that chromosome, a condition referred to as trisomy.
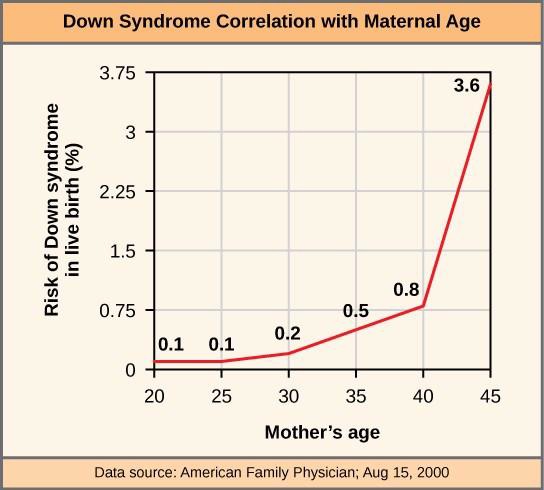
Monosomic human zygotes missing any one copy of an autosome invariably fail to develop to birth because they lack essential genes. Most autosomal trisomies also fail to develop to birth; however, trisomy of some of the smaller chromosomes (13, 15, 18, 21, or 22) can result in offspring that survive for several weeks to many years. Trisomic individuals suffer from an excess in gene dose, as they may synthesize too much of the gene products encoded by that chromosome. This extra dose of specific genes can lead to a number of functional challenges and often precludes development. The most common trisomy among viable births is that of chromosome 21, which corresponds to Down Syndrome. Individuals with this inherited disorder are characterized by short stature and stunted digits, facial distinctions that include a broad skull and large tongue, and significant developmental delays. The incidence of Down syndrome is correlated with maternal age; older women are more likely to become pregnant with fetuses carrying the trisomy 21 genotype (Figure 15.11).
Polyploidy
An individual with more than the correct number of chromosome sets is called polyploid. For instance, fertilization of an abnormal diploid egg with a normal haploid sperm would yield a triploid zygote. Polyploid animals are extremely rare and are usually sterile because meiosis cannot proceed normally. Rarely, polyploid animals can reproduce asexually. In contrast, polyploidy is very common in the plant kingdom, and polyploid plants tend to be larger and more robust than euploids of their species (Figure 15.12).

15.2.3 Sex Chromosome Nondisjunction in Humans
In contrast to autosomal trisomies and monosomies, humans can often function normally with different numbers of the X chromosome. In part, this occurs because of a process called X inactivation. Early in development, when female mammalian embryos consist of just a few thousand cells (relative to trillions in the newborn), one X chromosome in each cell inactivates by tightly condensing into a dormant structure called a Barr body. The chance that an X chromosome (maternally or paternally derived) is inactivated in each cell is random, but once the inactivation occurs, all cells derived from that one will have the same inactive X chromosome. By this process, females compensate for their double genetic dose of X chromosome. In so-called “tortoiseshell” cats, embryonic X inactivation is observed as color variegation (Figure 12.13). Females that are heterozygous for an X-linked coat color gene will express one of two different coat colors over different regions of their body, corresponding to whichever X chromosome is inactivated in the embryonic cell progenitor of that region.

X Chromosome Aneuploidy in Humans
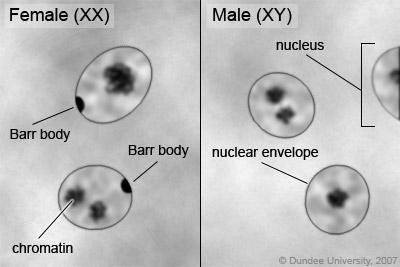
An individual carrying an abnormal number of X chromosomes will inactivate all but one X chromosome in each of her cells (Figure 15.14). However, even inactivated X chromosomes continue to express a few genes, and X chromosomes must reactivate for the proper maturation of female ovaries. As a result, X- chromosomal abnormalities are typically associated with mild mental and physical defects, as well as sterility.
Several errors in sex chromosome number have been characterized. Individuals with three X chromosomes (XXX) are phenotypically female but express developmental delays and reduced fertility. The XXY genotype, corresponding to one type of Klinefelter syndrome, corresponds to phenotypically male individuals with small testes, enlarged breasts, and reduced body hair. More complex types of Klinefelter syndrome exist in which the individual has as many as five X chromosomes. In all types, every X chromosome except one undergoes inactivation to compensate for the excess genetic dosage. This can be seen as several Barr bodies in each cell nucleus. Turner syndrome, characterized as an X0 genotype (i.e., only a single sex chromosome), corresponds to a phenotypically female individual with short stature, webbed skin in the neck region, hearing and cardiac impairments, and sterility.
Concept check
Do women with Turner syndrome have Barr bodies?
15.2.4 Duplications and Deletions
In addition to the loss or gain of an entire chromosome, a chromosomal segment may be duplicated or lost. Duplications and deletions often produce offspring that survive but exhibit physical and mental abnormalities. Duplicated chromosomal segments may fuse to existing chromosomes or may be free in the nucleus. Cri-du-chat (from the French for “cry of the cat”) is a syndrome associated with nervous system abnormalities and identifiable physical features that result from a deletion of most of the small arm of chromosome 5 (Figure 15.15). Infants with this genotype emit a characteristic high-pitched cry on which the disorder’s name is based.

15.2.5 Chromosomal Structural Rearrangements
Cytologists have characterized numerous structural rearrangements in chromosomes, but chromosome inversions and translocations are the most common.
Chromosome Inversions
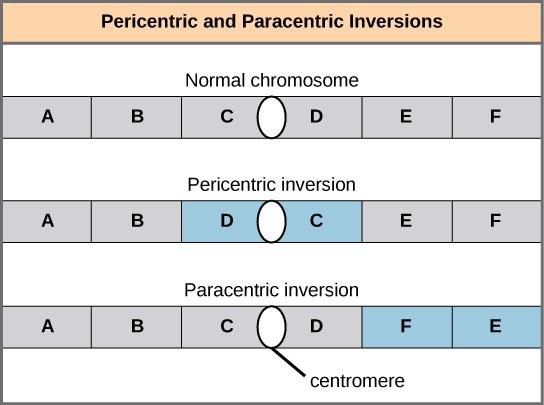
A chromosome inversion is the detachment, 180° rotation, and reinsertion of part of a chromosome. Unless they disrupt a gene sequence, inversions only change the orientation of genes and are likely to have mild effects. However, altered gene orientation can result in functional changes because regulators of gene expression could be moved out of position with respect to their targets, causing aberrant levels of gene products.
An inversion can be pericentric and include the centromere, or paracentric and occur outside of the centromere (Figure 15.16). A pericentric inversion that is asymmetric about the centromere can change the relative lengths of the chromosome arms, making these inversions easily identifiable.
Translocations
A translocation occurs when a segment of a chromosome dissociates and reattaches to a different chromosome. Translocations can be benign or have devastating effects depending on how the positions of genes are altered with respect to regulatory sequences. Notably, specific translocations have been associated with several cancers and with schizophrenia. Reciprocal translocations result from the exchange of chromosome segments between two nonhomologous chromosomes such that there is no gain or loss of genetic information (Figure 15.17).
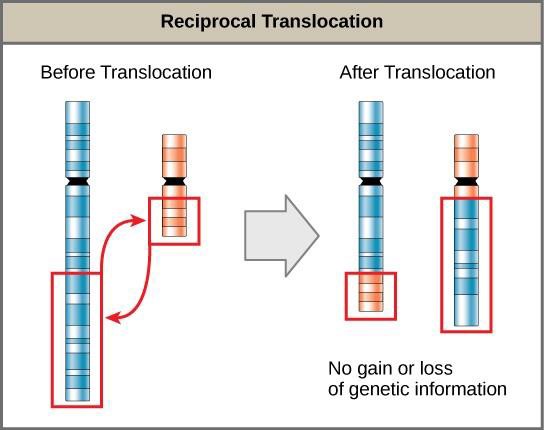
The Chromosome 18 Inversion
Not all structural rearrangements of chromosomes produce noviable, impaired, or infertile individuals. In rare instances, such a change can result in the evolution of a new species. In fact, a pericentric inversion in chromosome 18 appears to have contributed to the evolution of humans. This inversion is not present in our closest genetic relatives, the chimpanzees. Humans and chimpanzees differ by pericentric inversions on several chromosomes and by the fusion of two separate chromosomes in chimpanzees that correspond to chromosome two in humans.
The pericentric chromosome 18 inversion if believed to have occurred in early humans following their divergence from a common ancestor with chimpanzees approximately five million years ago. Researchers characterizing this inversion have suggested that approximately 19,000 nucleotide bases were duplicated on 18p, and the duplicated region inverted and reinserted on chromosome 18 of an ancestral human.
A comparison of human and chimpanzee genes in the region of this inversion indicates that two genes – ROCK1 and USP14 – that are adjacent on chimpanzee chromosome 17 (which corresponds to human chromosome 18) are more distantly positioned on human chromosome 18. This suggests that one of the inversion breakpoints occurred between these two genes. Interestingly, humans and chimpanzees express USP14 at distinct levels in specific cell types, including cortical cells and fibroblasts. Perhaps the chromosome 18 inversion in ancestral humans repositioned specific genes and reset their expression levels in a useful way. Because both ROCK1 and USP14 encode cellular enzymes, a change in their expression could alter cellular function. It is not known how this inversion contributed to hominid evolution, but it appears to be a significant factor in the divergence of humans from other primates.[1]
15.3 | Sexual Reproduction
Learning Objectives
By the end of this section, you will be able to:
- Explain that meiosis and sexual reproduction are evolved traits.
- Identify variation among offspring as a potential evolutionary advantage to sexual reproduction.
- Describe the three different life-cycle types among sexual multicellular organisms and their commonalities.
Sexual reproduction was an early evolutionary innovation after the appearance of eukaryotic cells. It appears to have been very successful because most eukaryotes are able to reproduce sexually, and in many animals, it is the only mode of reproduction. And yet, scientists recognize some real disadvantages to sexual reproduction.
On the surface, creating offspring that are genetic clones of the parent appears to be a better system. If the parent organism is successfully occupying a habitat, offspring with the same traits would be similarly successful. There is also the obvious benefit of not requiring another organism of the opposite sex. Indeed, some organisms that lead a solitary lifestyle have retained the ability to reproduce asexually. In addition, in asexual populations, every individual is capable of reproduction. Since males in sexual populations d o not p r o d u c e offspring, in theory an asexual population could grow twice as fast.
However, multicellular organisms that exclusively depend on asexual reproduction are exceedingly rare. Why is sexuality (and meiosis) so common? This is one of the important unanswered questions in biology and has been the focus of much research beginning in the latter half of the twentieth century. There are several possible explanations, one of which is that the variation that sexual reproduction creates among offspring is very important to the survival and reproduction of the population. The only source of variation in asexual organisms is mutation. This is the ultimate source of variation in sexual organisms, but in addition, those different mutations are continually reshuffled from one generation to the next when different parents combine their unique genomes and the genes are mixed into different combinations by crossovers during prophase I and random assortment at metaphase I.
15.3.1 Life Cycles of Sexually Reproducing Organisms
Fertilization and meiosis alternate in sexual life cycles. There are three main categories of life cycles in multicellular organisms: diploid-dominant, haploid-dominant, and alternation of generation.
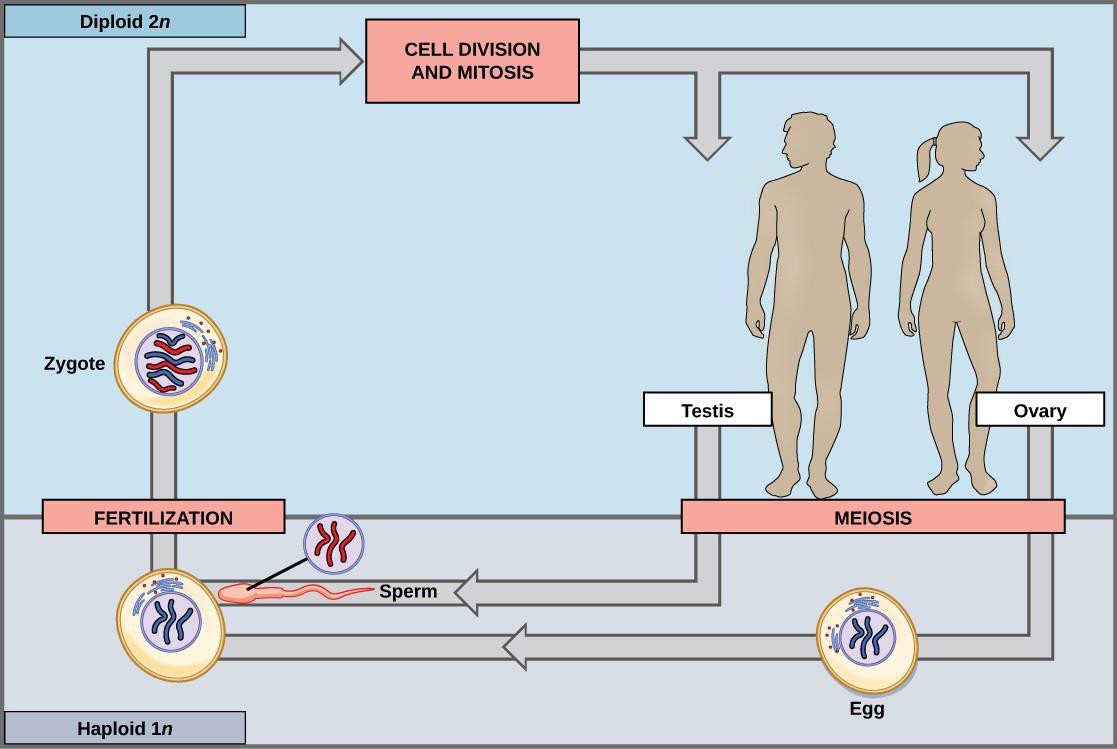
In diploid-dominant organisms, including most animals, the multicellular diploid stage is the most obvious life stage. In animals, the only haploid cells are the gametes. Early in the development of the embryo, specialized diploid cells, called germ cells, are produced within the gonads, such as the testes and ovaries. Germ cells are capable of mitosis to perpetuate the cell line and meiosis to produce gametes. Once the haploid gametes are formed, they lose the ability to divide again. There is no multicellular haploid life stage. Fertilization occurs with the fusion of two gametes, usually from different individuals, restoring the diploid state (Figure 15.2).
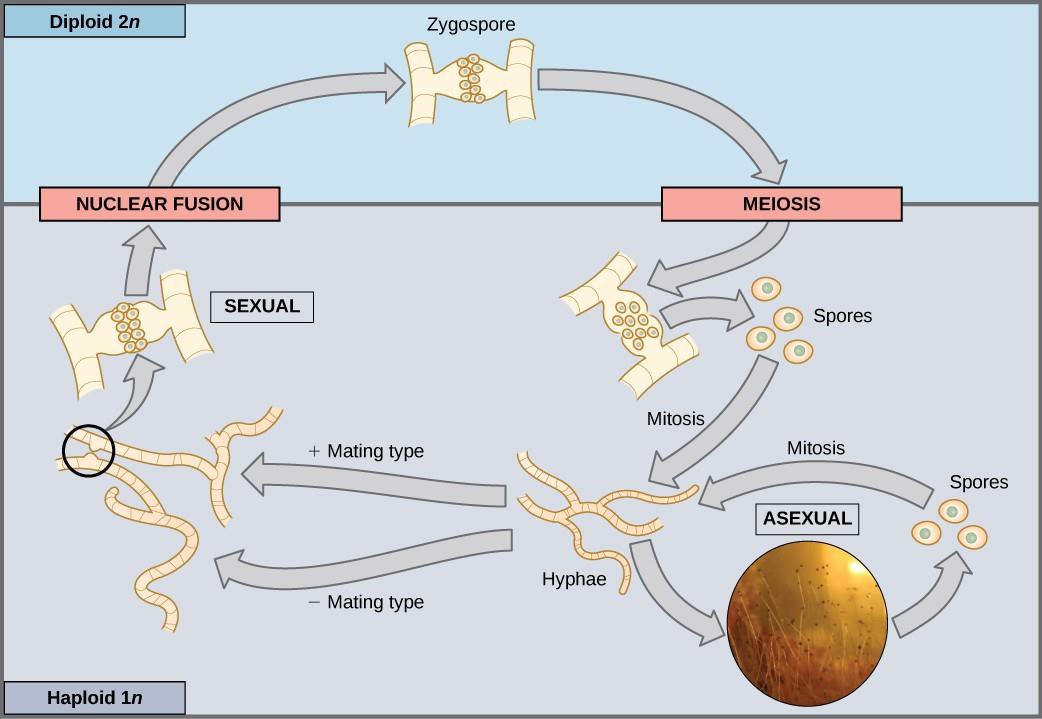
In haploid-dominant organisms, including fungi and some algae, the multicellular haploid stage is the most obvious life stage. The haploid cells that make up the tissues of the dominant multicellular stage are formed by mitosis. During sexual reproduction, specialized haploid cells from two individuals, designated the (+) and (−) mating types, join to form a diploid zygote. The zygote immediately undergoes meiosis to form four haploid cells called spores. Although haploid like the “parents,” these spores contain a new genetic combination from two parents. The spores can remain dormant for various time periods. Eventually, when conditions are conducive, the spores form multicellular haploid structures by many rounds of mitosis (Figure 15.18).
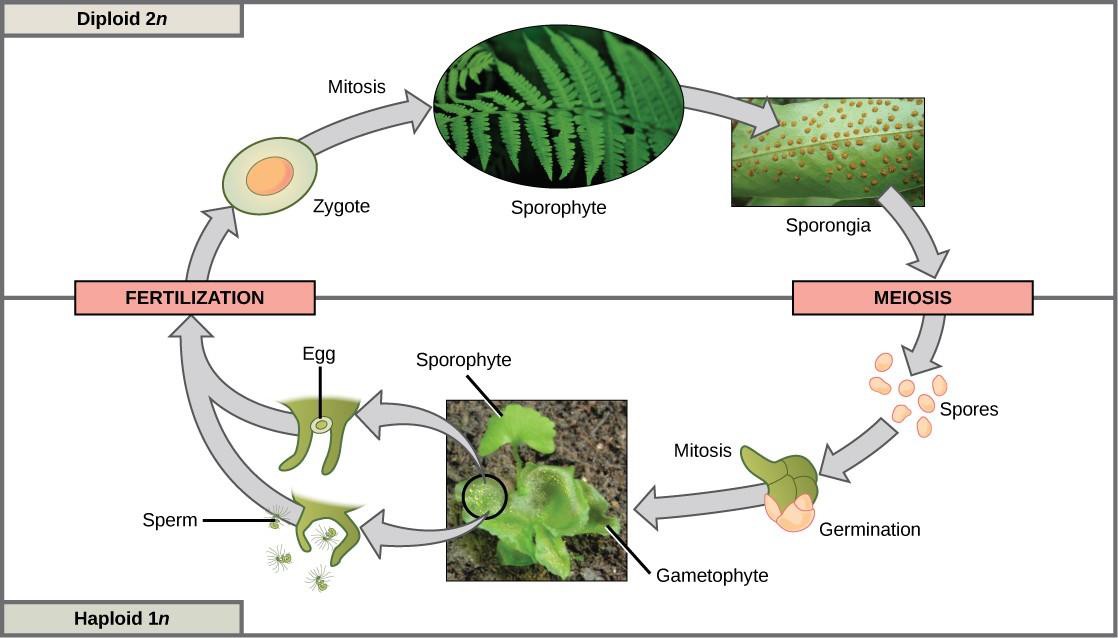
Organisms that show alternation of generations, including plants and some algae, have both haploid and diploid multicellular organisms as part of their life cycle. The haploid multicellular plants are called gametophytes, because they produce gametes from specialized cells. Meiosis is not directly involved in the production of gametes in this case, because the organism that produces the gametes is already a haploid. Fertilization between the gametes forms a diploid zygote. The zygote will undergo many rounds of mitosis and give rise to a diploid multicellular plant called a sporophyte. Specialized cells of the sporophyte will undergo meiosis and produce haploid spores. The spores will subsequently develop into the gametophytes (Figure 15.19).
In summary, sexual reproduction takes many forms in multicellular organisms. However, at some point in each type of life cycle, meiosis produces haploid cells that will fuse with the haploid cell of another organism. The mechanisms of variation—crossover, random assortment of homologous chromosomes, and random fertilization—are present in all versions of sexual reproduction. The fact that nearly every multicellular organism on Earth employs sexual reproduction is strong evidence for the benefits of producing offspring with unique gene combinations, though there are other possible benefits as well.

The Red Queen Hypothesis
It is not in dispute that sexual reproduction provides evolutionary advantages to organisms that employ this mechanism to produce offspring. But why, even in the face of fairly stable conditions, does sexual reproduction persist when it is more difficult and costly for individual organisms? Variation is the outcome of sexual reproduction, but why are ongoing variations necessary? Enter the Red Queen hypothesis, first proposed by Leigh Van Valen in 1973. The concept was named in reference to the Red Queen’s race inLewis Carroll’s book, Through the Looking-Glass.
All species co-evolve with other organisms; for example predators evolve with their prey, and parasites evolve with their hosts. Each tiny advantage gained by favorable variation gives a species an edge over close competitors, predators, parasites, or even prey. The only method that will allow a co-evolving species to maintain its own share of the resources is to also continually improve its fitness. As one species gains an advantage, this increases selection on the other species; they must also develop an advantage or they will be outcompeted. No single species progresses too far ahead because genetic variation among the progeny of sexual reproduction provides all species with a mechanism to improve rapidly. Species that cannot keep up become extinct. The Red Queen’s catchphrase was, “It takes all the running you can do to stay in the same place.” This is an apt description of co-evolution between competing species.
- Violaine Goidts et al., "Segment duplication associated with the human-specific inversion of chromosome 18: a further example of the impact of segmental duplications on karyotype and genome evolution in primates," Human Genetics. 115 (2004): 116-122. ↵

