1 Chapter 16. The Central Dogma: Genes to Traits
Chapter Outline
- 16.1 The Genetic Code
- 16.2 Transcription
- 16.3 RNA Processing in Eukaryotes
- 16.4 Translation
Introduction
Proteins are responsible for orchestrating nearly every cell function. Therefore, proteins also influence most of the traits displayed by multicellular organisms. So, how do cells know how to make the hundreds or thousands of different proteins that they need?
DNA contained in cells is a blueprint for making proteins (Figure 16.1). Each chromosome is a single molecule of DNA wound tightly around proteins. Segments of the DNA molecule, called genes, code for the order of amino acids in each protein. Both genes and the proteins they encode are absolutely essential to life as we know it.
16.1 | The Genetic Code
Learning Objectives
By the end of this section, you will be able to:
- Explain the Central Dogma of molecular biology.
- Describe the genetic code.
- Explain how the nucleotide sequence of a gene prescribes the amino acid sequence of a protein.
- Identify various types of point mutations and chromosomal mutations.
DNA contains the genes that serve as the blueprint for making proteins. Each protein has a unique sequence of amino acids. Since different amino acids have different chemistries (such as acidic vs. basic, or polar vs. nonpolar), the order of amino acids in a protein determines how the protein folds, thereby determining the shape and function of the protein. The order of amino acids in a protein is coded for by the gene for that protein.
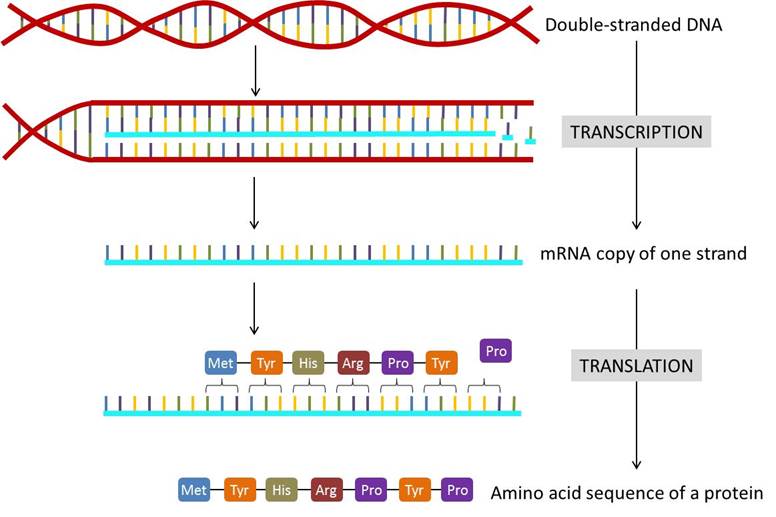
16.1 The Central Dogma: DNA Encodes mRNA and mRNA Encodes Protein
The flow of genetic information in cells from DNA to mRNA to protein is described by the Central Dogma of molecular biology (Figure 16.2). When a cell needs a particular protein, the gene that codes for that protein is activated and a single-stranded mRNA copy is made of the gene, in a process called transcription. The code copied into the mRNA is then used to determine the order of amino acids in the protein, in a process called translation. The copying of DNA to RNA is relatively straightforward, with one nucleotide being added to the mRNA strand for every nucleotide read in the DNA strand. The translation to protein is a bit more complex because three mRNA nucleotides correspond to one amino acid in the polypeptide sequence (Figure 16.2).
The Genetic Code Is Universal and Redundant
How does the order of nucleotides in an mRNA specify the order of amino acids in a protein? mRNA is “read” in three nucleotide segments called codons. Since RNA has four nucleotides (A, C, U, and G), there are 64 (43) possible combinations of three nucleotides (Figure 16.3). 61 of these codons code for one of the 20 common amino acids. The other three are called stop codons or nonsense codons because they do not code for an amino acid.
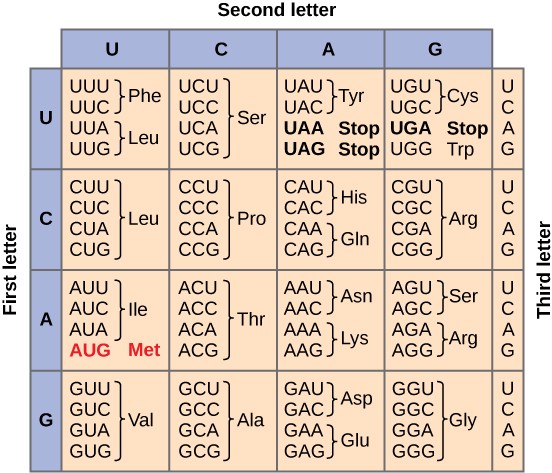
Scientists painstakingly solved the genetic code by translating synthetic mRNAs in vitro and sequencing the proteins they specified (Figure 16.4). Once all of the codons were known, they discovered some important features of the code.
The genetic code is universal. With a few exceptions, virtually all species use the same genetic code for protein synthesis. Conservation of codons means that a purified mRNA encoding the globin protein in horses could be transferred to a tulip cell, and the tulip would synthesize horse globin. That there is only one genetic code is powerful evidence that all of life on Earth shares a common origin
Since there are more nucleotide triplets than there are amino acids, the genetic code is redundant. In other words, a given amino acid can be encoded by more than one nucleotide triplet. Redundancy reduces the negative impact of random mutations. Codons that specify the same amino acid typically only differ by one nucleotide, usually the third one. For example, ACU, ACC, ACA and ACG all code for the amino acid threonine. In addition, amino acids with chemically similar side chains are encoded by similar codons. For example, UGU and UGC code for the amino acid cysteine, while AGU and AGC code for the amino acid serine. Cysteine and serine both have polar side chains that are very similar in size and other properties. Thus, the redundancy of the genetic code ensures that a single- nucleotide substitution mutation might specify either the same amino acid or a similar amino acid, preventing the protein from being rendered completely nonfunctional.
While 61 of the 64 codons specify the addition of a specific amino acid to a polypeptide chain, the remaining three codons terminate protein synthesis and release the polypeptide from the translation machinery. These triplets are called nonsense codons, or stop codons. Another codon, AUG, also has a special function. In addition to specifying the amino acid methionine, it also serves as the start codon to initiate translation. The reading frame for translation is set by the AUG start codon near the 5′ end of the mRNA.
Elucidating the Genetic Code
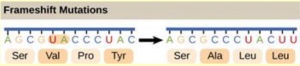
Given the different numbers of “letters” in the mRNA and protein “alphabets,” scientists theorized that combinations of nucleotides corresponded to single amino acids. Nucleotide doublets would not be sufficient to specify every amino acid because there are only 16 possible two-nucleotide combinations (42). In contrast, there are 64 possible nucleotide triplets (43). The fact that amino acids were encoded by nucleotide triplets was confirmed experimentally by Francis Crick and Sydney Brenner. They inserted one, two, or three nucleotides into the gene of a virus. When one or two nucleotides were inserted, the protein was not made. When three nucleotides were inserted, the protein was synthesized and functional. This demonstrated that three nucleotides specify each amino acid.The nucleotide triplets that code for amino acids are called codons. The insertion of one or two nucleotides completely changed the triplet reading frame, thereby altering the message for every subsequent amino acid (Figure 16.4). Though insertion of three nucleotides caused an extra amino acid to be inserted during translation, the integrity of the rest of the protein was maintained.
Exceptions to the Central Dogma
Many genes code for RNA molecules that do not function as mRNAs and are therefore not translated into proteins. Some RNAs, called rRNA, form parts of the ribosomes. Others form transfer RNAs, or tRNA, which help with translation. Still others can regulate which genes are expressed.
Another exception to the central dogma is in some cases, information flows backwards as is seen in certain viruses called retroviruses. These viruses have genes made up of RNA and when retroviruses infect a cell, the virus has to synthesize a DNA version of the RNA genes using a specialized viral polymerase called reverse transcriptase. The human immunodeficiency virus (HIV), which causes AIDS, is a retrovirus and many of the prescribed drugs used for AIDS patients target the HIV reverse transcriptase.
16.2 Mutations
In the living cell, DNA undergoes frequent chemical changes, especially as it is being replicated. These changes, result from incorrect nucleotides being inserted into the coding region of a gene. Most of these changes are quickly repaired. Those that are not repaired result in a mutation: a heritable change in the DNA.
Point Mutations
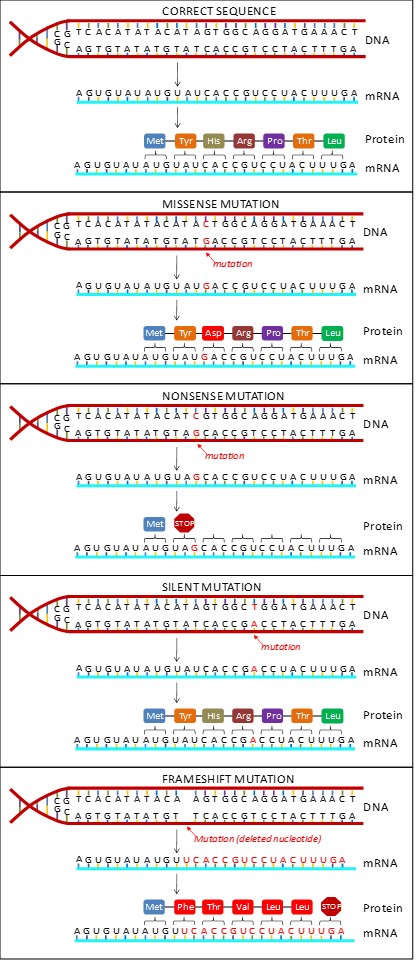
If a single base is changed in the DNA sequence, say from an A to a G, it results in a point mutation. A point mutation can have drastic consequences at the protein level depending on which base was substituted. Different types of point mutations are named based on their effect on the resulting protein. (Figure 16.5)
Missense Mutation
A missense mutation causes a different amino acid to be inserted in the protein. Even a single incorrect amino acid can have dire effects on an organism. For example, sickle-cell disease in humans results from the replacement of A by T at the 17th nucleotide of a hemoglobin gene, thereby substituting valine (GTG) for glutamic acid (GAG).
Nonsense Mutation
A nonsense mutation changes a codon that specified an amino acid to a stop codon. Translation of the resulting mRNA will therefore stop prematurely. The earlier in the gene that this mutation occurs, the shorter the protein will be and the more likely that it will be unable to function.
Silent Mutation
A silent mutation results in changing a codon to another codon that encodes the same amino acid. This is possible because the genetic code is redundant. Since this type of mutation does not change the protein product, it cannot be detected unless the gene is sequenced.
Frameshift Mutation
A frameshift mutation results when extra base pairs are added (insertion) or removed (deletion) from a gene. The resulting shift in the reading frame can have devastating consequences (Figure 16.4). Frameshifts can alter every subsequent codon and may also create stop codons.
Chromosome Mutations
Chromosomal mutations are large-scale mutations resulting in changes at the chromosome level instead of at the gene level. For example, a deletion results in a large section of a chromosome, usually involving several to hundreds of genes, being lost. A duplication is the opposite situation: a large section of a chromosome is repeated. In an inversion, a piece of a chromosome breaks off, flips, and rejoins the chromosome. A translocation results when a large segment of one chromosome breaks off and attaches to a different chromosome (Figure 16.6).
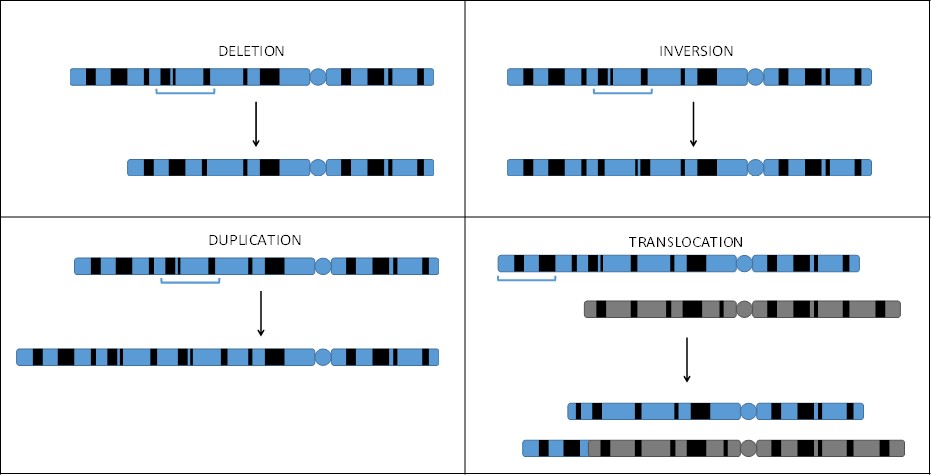
Since they involve large numbers of genes being removed or repeated, deletions and duplications often cause serious problems for the individual who has them. Conversely, inversions and translocations are often not problematic. Notice that in both cases, no genetic material is lost (Figure 5.6). The only problems that may arise are if the breakpoints are in the middle of genes. However, both inversions and translocations cause problems during reproduction and may lead to infertility.
Results of Mutations
Regardless of type, all mutations can be divided into three categories:
Beneficial mutations increase the fitness (its ability to survive and reproduce) of an organism.
Neutral mutations do not have an effect on fitness. Silent mutations fall in this category.
Deleterious mutations lower fitness in an organism.
16.2 | Transcription
Learning Objectives
By the end of this section, you will be able to:
- Describe the different steps in transcription.
- Discuss the role of promoters in transcription.
- Describe the role of RNA polymerases in transcription.
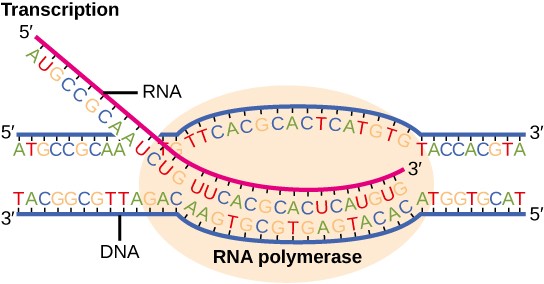
Transcription is the process of making an RNA copy of a gene (Figure 16.2). In eukaryotic cells, DNA is housed in the nucleus and transcription takes place in the nucleus. Since prokaryotic cells do not have a nucleus, transcription takes place in the cytoplasm in these cells.
Transcription is mediated by enzymes called RNA polymerases. RNA polymerases synthesize RNA in a 5′ ➝ 3′ direction. One of the strands of DNA, called the template strand, is used as a template to construct a complementary copy of the DNA. The other strand of DNA is nearly identical to the RNA copy, and is therefore called the coding strand. Since RNA has the nucleotide uracil (U) rather than thymine (T), the RNA copy has the same sequence as the coding strand except with U instead of T. For example, if the template strand of DNA has the sequence: ATCAGT, the coding strand will have the sequence TAGTCA and the RNA will have the sequence UAGUCA.
The first nucleotide pair in the DNA double helix, from which the first RNA nucleotide is transcribed is called the +1 site, or the initiation site. Nucleotides preceding the initiation site are given negative numbers and are designated upstream. Conversely, nucleotides following the initiation site are given positive numbers and are called downstream.
Transcription takes place in three stages: initiation, elongation, and termination.
16.2.1 Initiation of Transcription
Promoters
The first step in transcription is to identify where to begin copying the DNA strand. Each gene has a specific sequence of DNA, called the promotor, which specifies where to begin transcription and denotes which DNA strand to copy. The promoter is the site where the transcription machinery binds and initiates transcription. In most cases, promoters exist upstream of the genes they regulate.
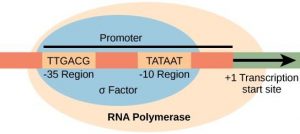
The specific sequence of a promoter is very important because it determines whether the corresponding gene is transcribed all the time, some of the time, or infrequently. Prokaryotic promoters have regions at -10 and -35 upstream of the initiation site, where the promoter binds (Figure 16.7). The -10 sequence is TATAAT, and is therefore called the TATA box. The -35 sequence is TTGACA. A subunit of RNA polymerase called sigma (σ) binds to the -35 sequence. Once this interaction is made, the rest of the RNA polymerase enzyme binds to the promoter. Eukaryotic promoters are much larger and more complex than prokaryotic promoters, but they also have a TATA box. (Figure 16.8).
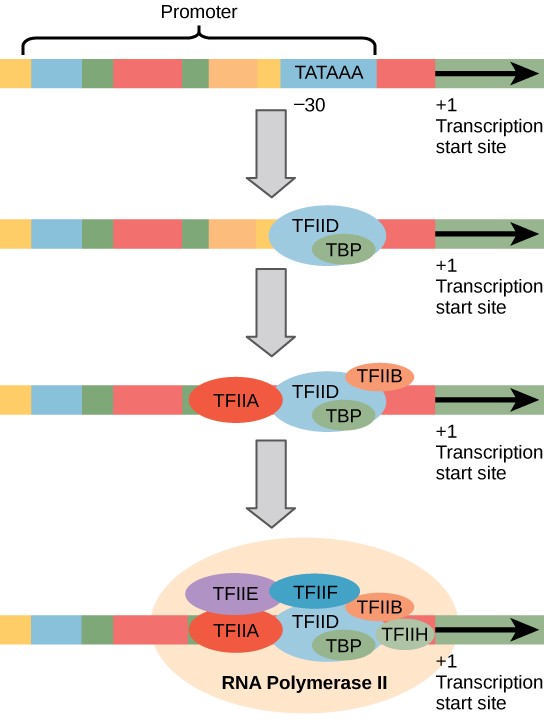
Initiation Complex
In prokaryotes, σ controls when RNA polymerase binds to a promoter and begins to transcribe a gene. Eukaryotes rely on an army of transcription factors and other regulatory proteins to help regulate the frequency with which RNA is synthesized from a gene. Transcription factors bind to the DNA template strand and subsequently recruit RNA polymerase for transcription initiation. Transcription factors are essential to ensure that the cell transcribes precisely the RNAs that it needs.
The complex of transcription factors and RNA polymerase bound to a promoter is called an initiation complex (Figure 16.8). Once RNA polymerase is bound to the promoter, the DNA double helix of a gene must be unwound to make room for RNA synthesis. The region of unwinding is called a transcription bubble (Figure 16.9).
16.2.2 Transcription Elongation
During elongation, RNA polymerase proceeds along the DNA template, synthesizing mRNA in the 5′ to 3′ direction at a rate of approximately 40 nucleotides per second. As elongation proceeds, the DNA is continuously unwound ahead of the enzyme and rewound behind it (Figure 16.9).
16.2.3 Transcription Termination
Once a gene is transcribed, the polymerase needs to dissociate from the DNA template and liberate the newly made mRNA. In prokaryotes, there are two kinds of termination signals. One is protein-based and the other is RNA-based.
Rho-dependent termination is controlled by the rho protein, which tracks along behind the polymerase on the growing mRNA chain. Near the end of the gene, the polymerase encounters a run of G nucleotides on the DNA template and it stalls. As a result, the rho protein collides with the polymerase. The interaction with rho releases the mRNA from the transcription bubble.
Rho-independent termination is controlled by specific sequences in the DNA template strand. As the polymerase nears the end of the gene being transcribed, it encounters a region rich in C–G nucleotides. The mRNA folds back on itself, and the complementary C–G nucleotides bind together. The result is a stable hairpin that causes the polymerase to break away and liberate the new mRNA transcript.
In eukaryotes, termination of transcription is different for the three different RNA polymerases. RNA Polymerase II transcribes 1,000–2,000 extra nucleotides beyond the end of the gene. This tail is subsequently removed during mRNA processing. Genes transcribed by RNA polymerase I contain a specific 18- nucleotide sequence that is recognized by a termination protein. The process of termination in RNA polymerase III involves an mRNA hairpin similar to rho-independent termination of transcription in prokaryotes.
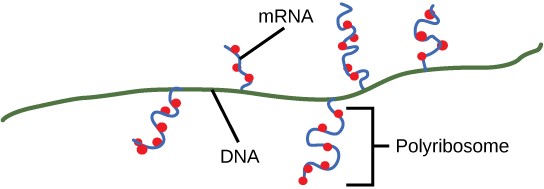
Upon termination, the process of transcription is complete. By the time termination occurs, the prokaryotic transcript would already have been used to begin synthesis of numerous copies of the encoded protein because these processes can occur concurrently (Figure 16.10). The unification of transcription and translation is possible because there is no nucleus in the prokaryotic cell. In contrast, the presence of a nucleus in eukaryotic cells precludes simultaneous transcription and translation.
Concept Check
A scientist splices a eukaryotic promoter in front of a bacterial gene and inserts the gene in a bacterial chromosome. Would you expect the bacteria to transcribe the gene?
RNA Polymerases
Prokaryotes use the same RNA polymerase to transcribe all of their genes. In E. coli, the polymerase is composed of five subunits, two of which are identical. Four of these subunits, denoted α, α, β, and β’ comprise the polymerase core enzyme. Each subunit has a unique role; the two α-subunits assemble the polymerase on the DNA; the β subunit binds to the ribonucleoside triphosphate that will become part of the new mRNA molecule; and the β’ binds the DNA template strand. The fifth subunit, σ, is involved only in transcription initiation. Without σ, the core enzyme would transcribe from random sites and would produce mRNA molecules that specified protein gibberish. The polymerase comprised of all five subunits is called the holoenzyme.
Eukaryotes employ three different RNA polymerases that each transcribe a different type of gene. Each RNA polymerase is made of 10 or more subunits. Each requires a distinct set of transcription factors to bring it to the DNA template.
RNA polymerase I transcribes most ribosomal RNA (rRNA), which becomes part of ribosomes. The rRNA molecules are considered structural RNAs because they are not translated into protein. The rRNAs are components of the ribosome and are essential to the process of translation.
RNA polymerase II synthesizes all protein-coding nuclear pre-mRNAs. Eukaryotic pre-mRNAs undergo extensive processing after transcription to become mature mRNAs. RNA polymerase II is responsible for transcribing the overwhelming majority of eukaryotic genes.
RNA polymerase III transcribes a variety of structural RNAs that includes one rRNA, transfer RNAs (tRNAs), and small nuclear RNAs (snRNAs). tRNAs have a critical role in translation; they bring amino acids to the growing polypeptide chain. snRNAs have a variety of functions, including “splicing” pre-mRNAs and regulating transcription factors.
A scientist characterizing a new gene can determine which polymerase transcribes it by testing whether the gene is expressed in the presence of a particular mushroom poison, α-amanitin. Interestingly, α-amanitin affects the three polymerases very differently. RNA polymerase I is completely insensitive to α-amanitin. In contrast, RNA polymerase II is extremely sensitive to α-amanitin, and RNA polymerase III is moderately sensitive. Knowing the transcribing polymerase can clue a researcher into the general function of the gene being studied.
16.3 | RNA Processing in Eukaryotes
Learning Objectives
By the end of this section, you will be able to:
- Describe the different steps in RNA processing.
- Understand the significance of exons, introns, and splicing.
- Explain how tRNAs and rRNAs are processed.
After transcription, eukaryotic pre-mRNAs must undergo several processing steps before they can be translated. Eukaryotic (and prokaryotic) tRNAs and rRNAs also undergo processing before they can function as components in the protein synthesis machinery.
16.3.1 mRNA Processing
The eukaryotic pre-mRNA undergoes extensive processing before it is ready to be translated. The additional steps involved in eukaryotic mRNA maturation create a molecule that is much more stable than a prokaryotic mRNA. Eukaryotic mRNAs last for several hours, whereas the typical E. coli mRNA lasts no more than five seconds.
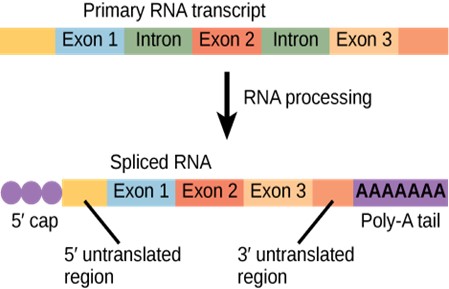
Pre-mRNAs are first coated in RNA-stabilizing proteins that protect the pre-mRNA from degradation while it is processed and exported out of the nucleus. The three most important steps of pre-mRNA processing are the addition of stabilizing and signaling factors at the 5′ and 3′ ends of the molecule, and the removal of intervening sequences that do not specify the appropriate amino acids.
5′ Capping
While the pre-mRNA is still being synthesized, a 7-methylguanosine cap is added to the 5′ end of the growing transcript. This “5’ cap” protects the mRNA from degradation. In addition, factors involved in protein synthesis recognize the cap to help initiate translation by ribosomes.
3′ Poly-A Tail
Once elongation is complete, the pre-mRNA is cleaved by an endonuclease between an AAUAAA consensus sequence and a GU-rich sequence, leaving the AAUAAA sequence on the pre-mRNA. An enzyme called poly-A polymerase then adds a string of approximately 200 A residues, called the poly-A tail. This modification further protects the pre-mRNA from degradation.
Pre-mRNA Splicing
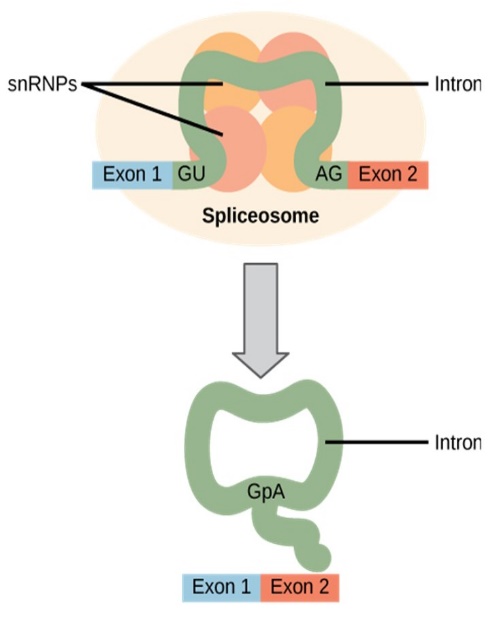
Eukaryotic genes are composed of exons, which correspond to protein-coding sequences (ex-on signifies that they are expressed), and intervening sequences called introns (intron denotes their intervening role). Intron sequences in mRNA do not encode functional proteins and are removed from the pre-mRNA during processing.
All of a pre-mRNA’s introns must be completely and precisely removed before protein synthesis begins. If the process errs by even a single nucleotide, the reading frame of the rejoined exons would shift, and the resulting protein would be dysfunctional (Figure 16.5). The process of removing introns and reconnecting exons is called splicing (Figure 16.11). Introns are removed and degraded while the pre-mRNA is still in the nucleus. Splicing occurs by a sequence-specific mechanism that ensures introns will be removed and exons rejoined with accuracy and precision. The splicing of pre-mRNAs is conducted by complexes of proteins and RNA molecules called spliceosomes. Note that more than 70 individual introns can be present, and each has to undergo the process of splicing – in addition to 5′ capping and the addition of a poly-A tail – just to generate a single, translatable mRNA molecule (Figure 16.12).
Concept Check
Errors in splicing are implicated in cancers and other human diseases. What kinds of mutations might lead to splicing errors? Think of different possible outcomes if splicing errors occur.
An Unexpected Discovery
The discovery of introns came as a surprise to researchers in the 1970s who expected that pre-mRNAs would specify protein sequences without further processing, as they had observed in prokaryotes. The genes of higher eukaryotes very often contain one or more introns. These regions may correspond to regulatory sequences; however, the biological significance of having many introns or having very long introns in a gene is unclear. It is possible that introns slow down gene expression because it takes longer to transcribe pre-mRNAs with lots of introns. Alternatively, introns may be nonfunctional sequence remnants left over from the fusion of ancient genes throughout evolution. This is supported by the fact that separate exons often encode separate protein subunits or domains. For the most part, the sequences of introns can be mutated without ultimately affecting the protein product.
16.3.2 Processing of tRNAs and rRNAs
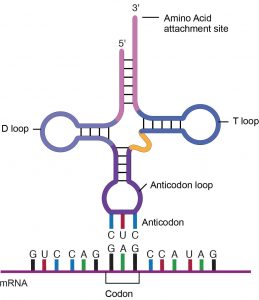
tRNAs and rRNAs are structural molecules that have roles in protein synthesis. However, these RNAs are not translated. Pre-rRNAs are transcribed, processed, and assembled into ribosomes, in a specialized region of the nucleus called the nucleolus. Ribosomes are described in the next section. Pre-tRNAs are transcribed and processed in the nucleus and then released into the cytoplasm. Mature tRNAs take on a three-dimensional structure through hydrogen bonding between nucleotides within the tRNA. The result is an amino acid binding site at one end of the tRNA and an anticodon at the other end (Figure 16.14). The anticodon is a three-nucleotide sequence in a tRNA that base pairs with a complementary mRNA codon.
16.4 | Ribosomes and Protein Synthesis
Learning Objectives
By the end of this section, you will be able to:
- Describe the different steps in protein synthesis.
- Discuss the role of ribosomes in protein synthesis.
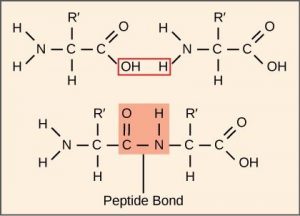
The synthesis of proteins consumes more of a cell’s energy than any other metabolic process. In turn, proteins account for more mass than any other component of living organisms (with the exception of water), and proteins perform virtually every function of a cell. The process of translation involves the decoding of an mRNA message to form a polypeptide. Fifty to more than 1000 amino acids are covalently strung together by dehydration synthesis reactions, forming peptide bonds. (Figure 16.15). This reaction is catalyzed by ribosomes.
16.4.1 The Protein Synthesis Machinery
In addition to the mRNA template, many molecules and macromolecules contribute to the process of translation. The composition of each component may vary across species; for instance, ribosomes may consist of different numbers of rRNAs and proteins. However, the general structures and functions of the protein synthesis machinery are comparable from bacteria to human cells. Translation requires the input of an mRNA template, ribosomes, tRNAs, and various enzymes.
Ribosomes
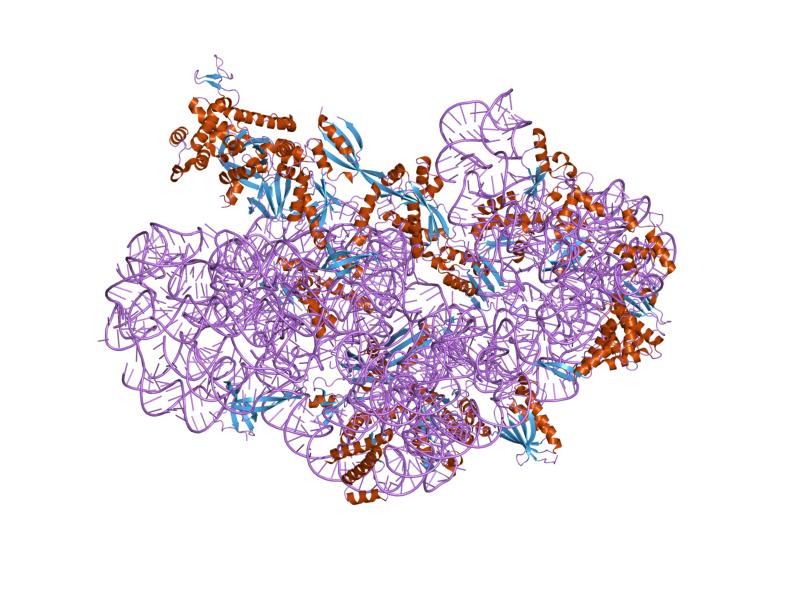
Before an mRNA is translated, a cell must build ribosomes. In a single prokaryotic E. coli cell, there are between 10,000 and 70,000 ribosomes present at any given time. A ribosome is a complex macromolecule composed of rRNAs and many distinct polypeptides (Figure 16.16). In eukaryotes, the nucleolus is specialized for the synthesis and assembly of ribosomes.
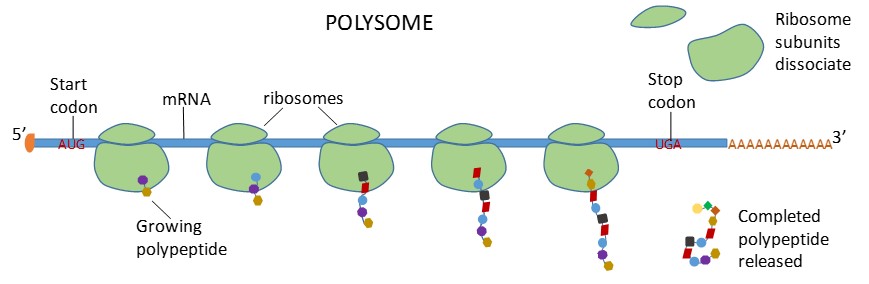
Ribosomes dissociate into large and small subunits when they are not synthesizing proteins and reassociate during the initiation of translation. The small subunit is responsible for binding the mRNA template, whereas the large subunit binds tRNAs. Each mRNA molecule is simultaneously translated by many ribosomes, all synthesizing protein in the same direction: reading the mRNA from 5′ to 3′ and synthesizing the polypeptide from the N terminus to the C terminus. The complete mRNA/poly-ribosome structure is called a polysome (Figure 16.17).
Ribosomes are found either in the cytoplasm or attached to the rough endoplasmic reticulum. Mitochondria and chloroplasts also have their own ribosomes, which look and act more similar to prokaryotic ribosomes than to the nuclear ribosomes in the same cell.
tRNAs
tRNAs are structural RNA molecules that bind to sequences on the mRNA template and add the corresponding amino acid to the polypeptide chain. Therefore, tRNAs are the molecules that actually “translate” the language of RNA into the language of proteins.
Each tRNA anticodon can base pair with one of the mRNA codons and add an amino acid or terminate translation, according to the genetic code. For instance the codon GAG on an mRNA template would bind a tRNA that had the complementary anticodon CUC and was linked to the amino acid glutamic acid (Figure 16.14).
Aminoacyl tRNA Synthetases
tRNA molecules are linked to their correct amino acids by a group of enzymes called aminoacyl tRNA synthetases. Adding the amino acid to a tRNA is called “charging” the tRNA. At least one type of aminoacyl tRNA synthetase exists for each of the 20 amino acids.
16.4.2 The Mechanism of Protein Synthesis
As with mRNA synthesis, protein synthesis can be divided into three phases: initiation, elongation, and termination.
Initiation of Translation
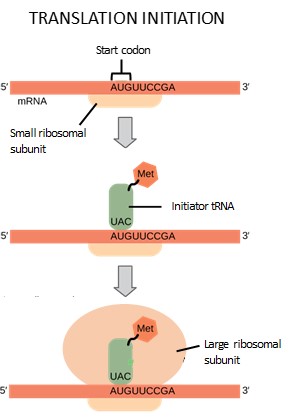
Protein synthesis begins with the formation of an initiation complex. This complex involves the small ribosomal subunit, the mRNA template, initiation factors, and a special initiator tRNA, called tRNAMet. The initiator tRNA has anticodon UAG, which interacts with the start codon AUG and is charged with the amino acid methionine. Methionine is therefore the first amino acid of every polypeptide chain.
First, the mRNA binds to the small ribosomal subunit. Initiation factors help the small subunit bind and scan along until it identifies the AUG start codon. Next, the anticodon of the initiator tRNA hydrogen bonds to the start codon. Finally, the large subunit of the ribosome binds, lining up so the initiator tRNA is in the P site. This step completes initiation of translation (Figure 16.18).
Elongation and Termination of Translation
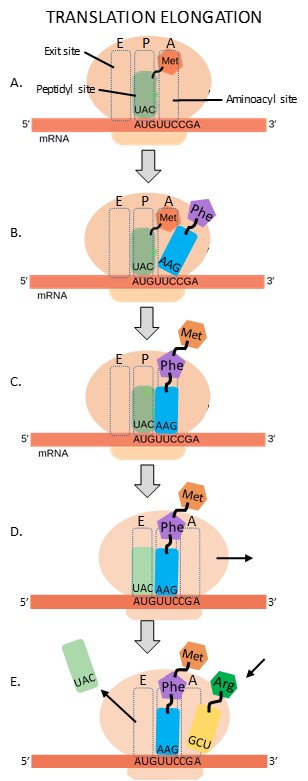
The large ribosomal subunit contains three tRNA binding sites: the A (aminoacyl) site, the P (peptidyl) site, and the E (exit) site (Figure 16.19A). During translation elongation, the mRNA template provides specificity. As the ribosome moves along the mRNA, each mRNA codon comes into register, and specific binding with the corresponding charged tRNA anticodon is ensured.
The first step in elongation occurs when the second codon is recognized by the appropriate charged tRNA. As the anticodon of this tRNA hydrogen bonds to the second codon, it enters the A site of the ribosome (Figure 16.19B).
Next, the two amino acids are covalently bonded together with a peptide bond. The growing polypeptide chain remains attached to the tRNA in the A site (Figure 16.19C).
The ribosome now slides along the mRNA by a distance of one codon, in a process called translocation. Since the tRNAs are hydrogen bonded to the mRNA, they remain behind as the ribosome moves. The result of translocation is that the initiator tRNA is now in the E site of the ribosome, the second tRNA is in the P site, and the A site is empty (Figure 16.19D).
After translocation, the initiator tRNA exits from the E site. The third tRNA recognizes the third codon and hydrogen bonds to it, entering the A site of the ribosome (Figure 16.19E).
Steps C, D and E repeat until the stop codon is reached. Each cycle of these three steps adds one amino acid to the growing polypeptide. Amazingly, the E. coli translation apparatus takes only 0.05 seconds to add each amino acid, meaning that a 200-amino acid protein can be translated in just 10 seconds!
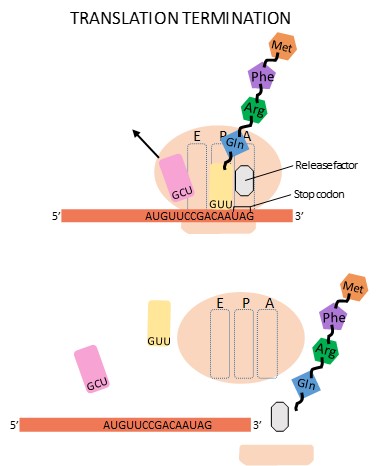
Termination of translation occurs when a stop codon (UAA, UAG, or UGA) is encountered. Upon aligning with the A site, the stop codon is recognized by a release factor. The P-site amino acid detaches from its tRNA, and the newly made polypeptide is released. The small and large ribosomal subunits dissociate from the mRNA and from each other and find another mRNA to translate (Figure 16.20). After many ribosomes have translated it, the mRNA is degraded so that its nucleotides can be reused.
16.4.3 Protein Folding, Modification, and Targeting
During and after translation, individual amino acids may be chemically modified, signal sequences may be added, and the new protein “folds” into a distinct three-dimensional structure as a result of intramolecular interactions. A signal sequence is a short tail of amino acids that directs a protein to a specific cellular compartment. These sequences at the amino end or the carboxyl end of the protein can be thought of as the protein’s “train ticket” to its ultimate destination. Other cellular factors recognize each signal sequence and help transport the protein from the cytoplasm to its correct compartment. For instance, a specific sequence at the amino terminus will direct a protein to the mitochondria or chloroplasts (in plants). Once the protein reaches its cellular destination, the signal sequence is usually clipped off.
Many proteins fold spontaneously, but some proteins require helper molecules, called chaperones, to help them fold correctly. Even if a protein is properly specified by its corresponding mRNA, it could take on a completely dysfunctional shape if abnormal temperature or pH conditions prevent it from folding correctly.

