Chapter 13. The Cell Cycle & Mitosis

Chapter Outline
- 13.1 DNA Organization and the Cell Cycle
- 13.2 The Cell Cycle
- 13.3 Control of the Cell Cycle
- 13.4 Cancer and the Cell Cycle
- 13.5 Prokaryotic Cell Division
Introduction
A human, as well as every sexually reproducing organism, begins life as a fertilized egg or zygote. Trillions of cell divisions subsequently occur in a controlled manner to produce a complex, multicellular human. In other words, that original single cell is the ancestor of every other cell in the body. Once a being is fully grown, cell reproduction is still necessary to repair or regenerate tissues. For example, new blood and skin cells are constantly being produced. All multicellular organisms use cell division for growth, maintenance, and repair of cells and tissues. Cell division is tightly regulated, and the occasional failure of regulation can have life-threatening consequences. Single-celled organisms use cell division as their method of reproduction.
13.1 | DNA Organization and the Cell Cycle
Learning Objectives
By the end of this section, you will be able to:
- Describe the structure of prokaryotic and eukaryotic genomes.
- Distinguish between chromosomes, genes, and traits.
- Describe the mechanisms of chromosome compaction.
The continuity of life from one cell to another has its foundation in the reproduction of cells by way of the cell cycle. The cell cycle is an orderly sequence of events that describes the stages of a cell’s life from the division of a single parent cell to the production of two genetically identical new daughter cells. The mechanisms involved in the cell cycle are highly regulated.
13.1.1 Genomic DNA
Before discussing the steps a cell must undertake to replicate, we need a deeper understanding of the structure and function of a cell’s genetic information. A cell’s DNA, packaged as double-stranded DNA molecules, is called its genome.
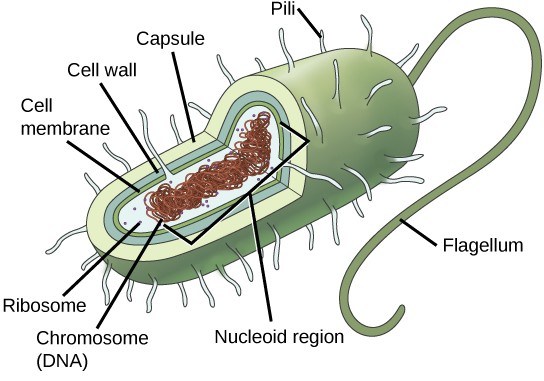
In prokaryotes, the genome is composed of a single circular double-stranded DNA molecule. (Figure 13.2). The region in the cell containing this genetic material is called a nucleoid. Some prokaryotes also have smaller loops of non- essential DNA called plasmids. Bacteria can exchange these plasmids with other bacteria, sometimes receiving beneficial new genes that the recipient can add to their chromosomal DNA. Antibiotic resistance is one trait that often spreads through a bacterial colony through plasmid exchange.
In eukaryotic cells, the genome consists of several double-stranded linear DNA molecules (Figure 13.3). Each species has a characteristic number of chromosomes in the nuclei of its cells. Human body cells have 46 chromosomes, while human gametes (sperm or eggs) have 23 chromosomes each. A typical body cell, or somatic cell, contains two matched sets of chromosomes, a configuration known as diploid. The letter n is used to represent a single set of chromosomes; therefore, a diploid organism is designated 2n. Human cells that contain one set of chromosomes are called gametes, or sex cells; these are eggs and sperm, and are designated 1n, or haploid.
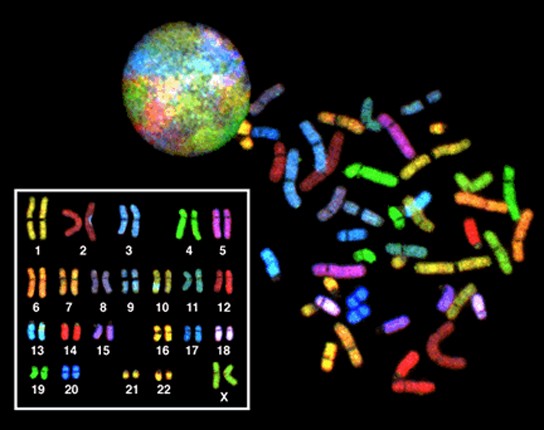
Matched pairs of chromosomes in a diploid organism are called homologous (“same knowledge”) chromosomes. Homologous chromosomes are the same length and have specific nucleotide segments called genes in exactly the same location, or locus. Genes are the functional units of chromosomes and determine specific characteristics by coding for specific proteins. Traits are the variations of those characteristics. For example, hair color is a characteristic with traits that are blonde, brown, or black. Each copy of a homologous pair of chromosomes originates from a different parent; therefore, the genes themselves are not identical. The variation of individuals within a species is due to the specific combination of the genes inherited from both parents. Even a slightly altered sequence of nucleotides within a gene can result in an alternative trait.
For example, there are three possible gene sequences on the human chromosome that code for blood type: sequence A, sequence B, and sequence O. Because all diploid human cells have only two copies of the chromosome that determines blood type, the blood type trait is determined by which two versions of the marker gene are inherited. It is possible to have two copies of the same gene sequence on both homologous chromosomes, with one on each (for example, AA, BB, or OO), or two different sequences, such as AO, BO, or AB. Minor variations of traits, such as blood type, eye color, and handedness all contribute to the natural variation found within a species. If the entire DNA sequence from any pair of human homologous chromosomes is compared, the difference is less than one percent. The sex chromosomes, X and Y, are the single exception to the rule of homologous chromosome uniformity: Other than a small amount of homology that is necessary to accurately produce gametes, the genes found on the X and Y chromosomes are different
13.1.2 Eukaryotic Chromosomal Structure and Compaction
If the DNA from all 46 chromosomes in a human cell nucleus was laid out end to end, it would measure approximately two meters; however, its diameter would be only 2 nm. Considering that the size of a typical human cell is about 10 µm (100,000 cells lined up to equal one meter), DNA must be tightly packaged to fit in the cell’s nucleus. At the same time, it must also be readily accessible for the genes to be expressed. During some stages of the cell cycle, the long strands of DNA are condensed into compact chromosomes. There are a number of ways that chromosomes are compacted.
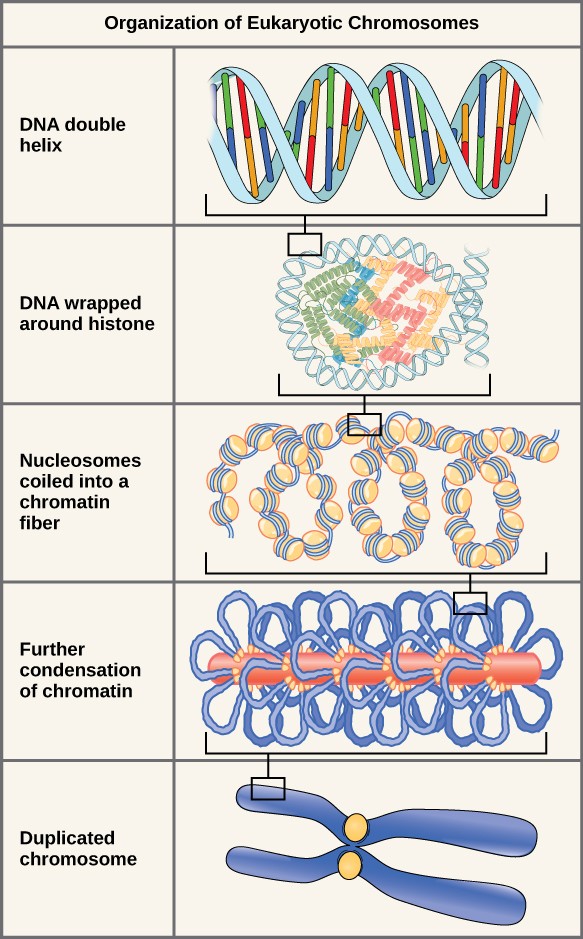
In the first level of compaction, short stretches of the DNA double helix wrap around a core of eight histone proteins at regular intervals along the entire length of the chromosome (Figure 13.4). The DNA-histone complex is called chromatin. The beadlike, histone DNA complex is called a nucleosome, and DNA connecting the nucleosomes is called linker DNA. A DNA molecule in this form is about seven times shorter than the double helix without the histones, and the beads are about 10 nm in diameter, in contrast with the 2-nm diameter of a DNA double helix. The next level of compaction occurs as the nucleosomes and the linker DNA between them are coiled into a 30-nm chromatin fiber. This coiling further shortens the chromosome so that it is about 50 times shorter. In the third level of packing, a variety of fibrous proteins is used to pack the chromatin. These fibrous proteins also ensure that each chromosome in a non-dividing cell occupies a particular area of the nucleus that does not overlap with that of any other chromosome (see the top image in Figure 13.4).
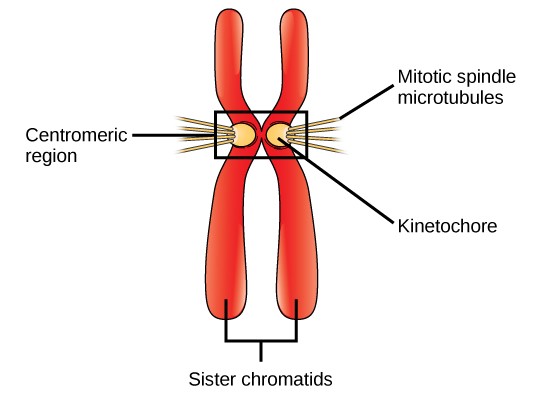
DNA replicates in the S phase of interphase. After replication, the chromosomes are composed of two linked sister chromatids. When fully compact, the pairs of identically packed chromosomes are bound to each other by cohesin proteins. The connection between the sister chromatids is closest in a region called the centromere. The conjoined sister chromatids, with a diameter of about 1 µm, are visible under a light microscope. The centromeric region is highly condensed and thus will appear as a constricted area.
13.2 | The Cell Cycle
Learning Objectives
By the end of this section, you will be able to:
- Describe the three stages of interphase.
- Discuss the behavior of chromosomes during mitosis.
- Explain how the cytoplasmic content is divided during cytokinesis.
- Define the quiescent G0 phase.
The cell cycle is an ordered series of events involving cell growth and cell division that produces two new daughter cells. Somatic cells on the path to cell division proceed through a series of precisely timed and carefully regulated stages of growth, DNA replication, and division that produces two genetically identical cells. In other words, a typical 2n somatic cell will divide into two 2n somatic cells that are genetically identical. This is a form of asexual reproduction.
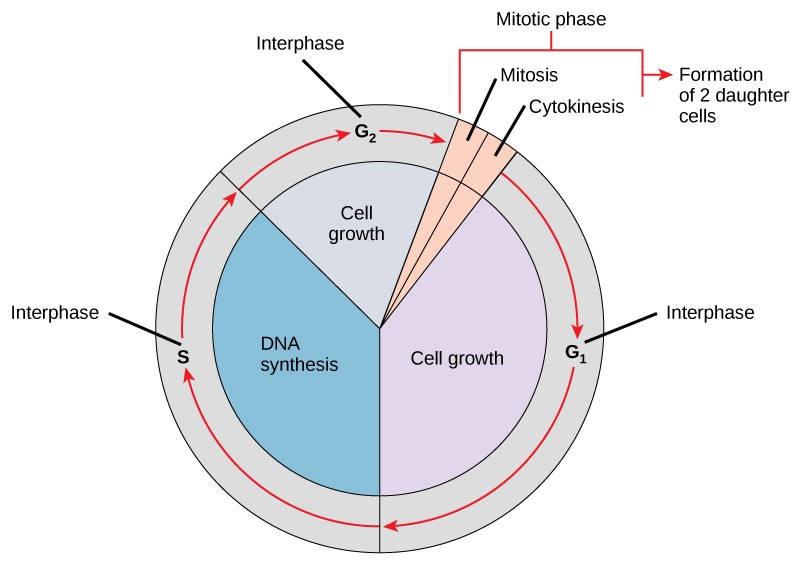
As a quick overview of the cell cycle, we see two major phases: interphase and the M phase (Figure 13.5). During interphase, the cell undergoes three distinct periods: G1, S, and G2. During G1, S, and G2, the cell grows, DNA is replicated, and the cell grows some more. During the M phase, the cell undergoes two distinct periods: mitosis (also called karyokinesis) and cytokinesis, the division of the cytoplasm.
13.2.1 Interphase
During interphase, the cell undergoes normal growth processes while also preparing for cell division. In order for a cell to move from interphase into the mitotic phase, many internal and external conditions must be met. The three aspects or stages of interphase are called G1, S, and G2.
G1 Phase (First Gap)
The first stage of interphase is called the G1 phase (first gap) because, from a microscopic aspect, little change is visible. However, during the G1 stage, the cell is quite active at the biochemical level. The cell is accumulating the building blocks of chromosomal DNA and the associated proteins as well as accumulating sufficient energy reserves to complete the task of replicating each chromosome in the nucleus.
S Phase (Synthesis of DNA)
Throughout interphase, nuclear DNA remains in a semi-condensed chromatin configuration. In the S phase, DNA replication can proceed through the mechanisms that result in the formation of identical pairs of DNA molecules—sister chromatids—that are firmly attached to the centromeric region. The centrosome is duplicated during the S phase. The two centrosomes will give rise to the mitotic spindle, the apparatus that orchestrates the movement of chromosomes during mitosis. At the center of each animal cell, the centrosomes of animal cells are associated with a pair of rod-like objects, the centrioles, which are at right angles to each other. Centrioles help organize cell division. Centrioles are not present in the centrosomes of other eukaryotic species, such as plants and most fungi.
G2 Phase (Second Gap)
In the G2 phase, the cell replenishes its energy stores and synthesizes proteins necessary for chromosome manipulation. Some cell organelles are duplicated, and the cytoskeleton is dismantled to provide resources for the mitotic phase. There may be additional cell growth during G2. The final preparations for the mitotic phase must be completed before the cell is able to enter the first stage of mitosis.
13.2.2 The Mitotic Phase
The mitotic phase is a multistep process during which the duplicated chromosomes are aligned, separated, and moved into two new, identical daughter cells. M phase is divided into mitosis and cytokinesis.
Mitosis
Mitosis is divided into a series of phases—Prophase, Prometaphase, Metaphase, Anaphase, and Telophase—that result in the division of the cell nucleus (Figure 13.6).
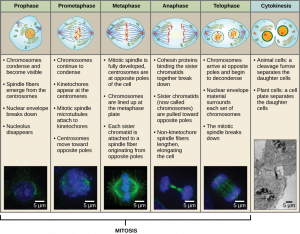
During Prophase, the nuclear envelope starts to dissociate into small vesicles and the membranous organelles (such as the Golgi complex or Golgi apparatus, and endoplasmic reticulum), fragment and disperse toward the periphery of the cell. The nucleolus disappears (disperses). The centrosomes begin to migrate to opposite poles of the cell. Microtubules that will form the mitotic spindle extend between the centrosomes, pushing them farther apart as the microtubule fibers lengthen. The sister chromatids begin to coil more tightly with the aid of condensin proteins and become visible under a light microscope.
During Prometaphase, many processes that were begun in prophase continue to advance. The remnants of the nuclear envelope fragment. The mitotic spindle continues to develop as more microtubules assemble and stretch across the length of the former nuclear area. Chromosomes become more condensed and discrete. Each sister chromatid develops a protein structure called a kinetochore in the centromeric region (Figure 13.7). The proteins of the kinetochore attract and bind mitotic spindle microtubules. As the spindle microtubules extend from the centrosomes, some of these microtubules come into contact with and firmly bind to the kinetochores. Once a mitotic fiber attaches to a chromosome, the chromosome will be oriented until the kinetochores of sister chromatids face the opposite poles. Eventually, all the sister chromatids will be attached via their kinetochores to microtubules from opposing poles. Spindle microtubules that do not engage the chromosomes are called polar microtubules. These microtubules overlap each other midway between the two poles and contribute to cell elongation. Astral microtubules are located near the poles, aid in spindle orientation, and are required for the regulation of mitosis.
During Metaphase, all the chromosomes are aligned in a plane called the metaphase plate, or the equatorial plane, midway between the two poles of the cell. The sister chromatids are still tightly attached to each other by cohesin proteins. At this time, the chromosomes are maximally condensed and are at their most visible under a microscope.
During Anaphase, the cohesin proteins degrade, and the sister chromatids separate at the centromere. Each chromatid, now called a chromosome, is pulled rapidly toward the centrosome to which its microtubule is attached. The cell becomes visibly elongated (oval shaped) as the polar microtubules slide against each other at the metaphase plate where they overlap.
During Telophase, the chromosomes reach the opposite poles, the nuclear envelopes reforms around the chromosomes. The mitotic spindles are depolymerized into tubulin monomers that will be used to assemble cytoskeletal components for each daughter cell. The enclosed chromosomes begin to decondense (unravel), relaxing into a chromatin configuration, and nucleosomes appear within the nuclear area. The cell will appear as a cell with two distinct nuclei. At this stage, mitosis is complete and division of the cytoplasm will follow during cytokinesis.
Cytokinesis
Cytokinesis, or “cell motion,” is the second main stage of the mitotic phase during which cell division is completed via the physical separation of the cytoplasmic components into two daughter cells. Division is not complete until the cell components have been apportioned and completely separated into the two daughter cells. Although the stages of mitosis are similar for most eukaryotes, the process of cytokinesis is quite different for eukaryotes that have cell walls, such as plant cells.
In cells such as animal cells that lack cell walls, cytokinesis actually begins at the midpoint of Anaphase. A contractile ring composed of actin filaments forms just inside the plasma membrane at the former Metaphase plate. The actin filaments pull the equator of the cell inward, forming a constriction. This constriction or fissure is called the cleavage furrow. The furrow deepens as the actin ring contracts, and eventually the membrane is cleaved in two (Figure 13.8).
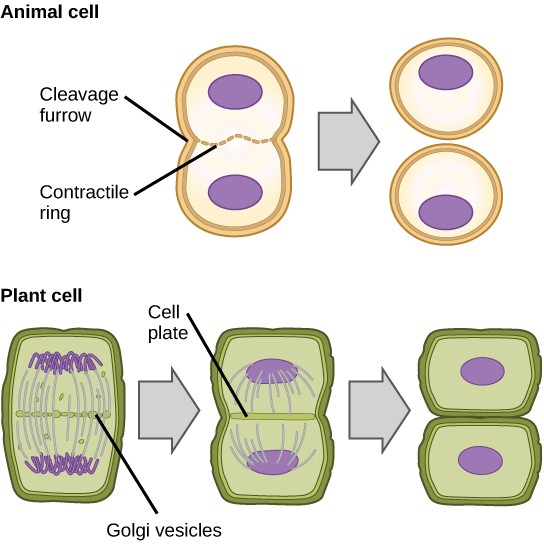
In plant cells, a new cell wall must form between the daughter cells. During interphase, the Golgi apparatus accumulates enzymes, structural proteins, and glucose molecules prior to breaking into vesicles and dispersing throughout the dividing cell. During telophase, these Golgi vesicles are transported on microtubules to form a phragmoplast (a vesicular structure) at the metaphase plate. There, the vesicles fuse and coalesce from the center toward the cell walls; this structure is called a cell plate.
As more vesicles fuse, the cell plate enlarges until it merges with the cell walls at the periphery of the cell. Enzymes use the glucose that has accumulated between the membrane layers to build a new cell wall made of cellulose. Remember that cellulose is a structural polymer of glucose. The Golgi membranes become parts of the plasma membrane on either side of the new cell wall (Figure 13.8).
13.2.3 G0 Phase (A variation of the cell cycle)
Not all cells adhere to the classic cell cycle pattern in which a newly formed daughter cell immediately enters the preparatory phases of interphase, closely followed by the mitotic phase. Cells in G0 phase (“G zero”) are not actively preparing to divide. The cell is in a quiescent (inactive) stage that occurs when cells exit the cell cycle. Some cells enter G0 temporarily until an external signal triggers the onset of G1. Other cells that never or rarely divide, such as mature cardiac muscle and nerve cells, remain in G0 permanently. Although cells in G0 are inactive in the sense that they are not actively preparing for and undergoing cell division, they can be very active in other ways. Many cells in the adult body for instance, are permanently in G0 while fulfilling their specialized functions.
13.3 | Control of the Cell Cycle
Learning Objectives
By the end of this section, you will be able to:
- Understand how the cell cycle is controlled by mechanisms both internal and external to the cell.
- Explain how the three internal control checkpoints occur at the end of G1, at the G2/M transition, and during metaphase.
- Describe the molecules that control the cell cycle through positive and negative regulation.
The length of the cell cycle is highly variable, even within the cells of a single organism. In humans, the frequency of cell turnover ranges from a few hours in early embryonic development, to an average of two to five days for epithelial cells. Some cells, such as cortical neurons or cardiac muscle cells spend all of their time in G0 and never divide. In early embryos of fruit flies, the cell cycle is completed in about eight minutes. The timing of events in the cell cycle is controlled by mechanisms that are both internal and external to the cell.
There is also variation in the time that a cell spends in each phase of the cell cycle. When fast-dividing mammalian cells are grown in culture (outside the body under optimal growing conditions), the length of the cycle is about 24 hours. In rapidly dividing human cells with a 24-hour cell cycle, the G1 phase lasts approximately 9 hours, the S phase lasts 10 hours, the G2 phase lasts about 4.5 hours, and the M phase lasts approximately 0.5 hours.
13.3.1 Regulation of the Cell Cycle by External Events
Both the initiation and inhibition of cell division are triggered by events external to the cell when it is about to begin the replication process. An event may be as simple as the death of a nearby cell or as sweeping as the release of growth- promoting hormones, such as human growth hormone (HGH). A lack of HGH can inhibit cell division, resulting in dwarfism, whereas too much HGH can result in gigantism. Crowding of cells can also inhibit cell division. Another factor that can initiate cell division is the size of the cell; as a cell grows, it becomes inefficient due to its decreasing surface-to-volume ratio. The solution to this problem is to divide.
Whatever the source of the message, the cell receives the signal, and a series of events within the cell allows it to proceed into interphase. Moving forward from this initiation point, every parameter required during each cell cycle phase must be met or the cycle cannot progress.
13.3.2 Regulation of the Cell Cycle at Internal Checkpoints
It is essential that the daughter cells that have been produced be exact duplicates of the parent cell. Mistakes in the duplication or distribution of the chromosomes lead to mutations that may be passed forward to every new cell produced from an abnormal cell. To prevent a compromised cell from continuing to divide, there are internal control mechanisms that operate at three main cell cycle checkpoints. A checkpoint is one of several points in the eukaryotic cell cycle at which the progression of a cell to the next stage in the cycle can be halted until conditions are favorable. These checkpoints occur near the end of G1, at the G2/M transition, and during metaphase (Figure 13.10).
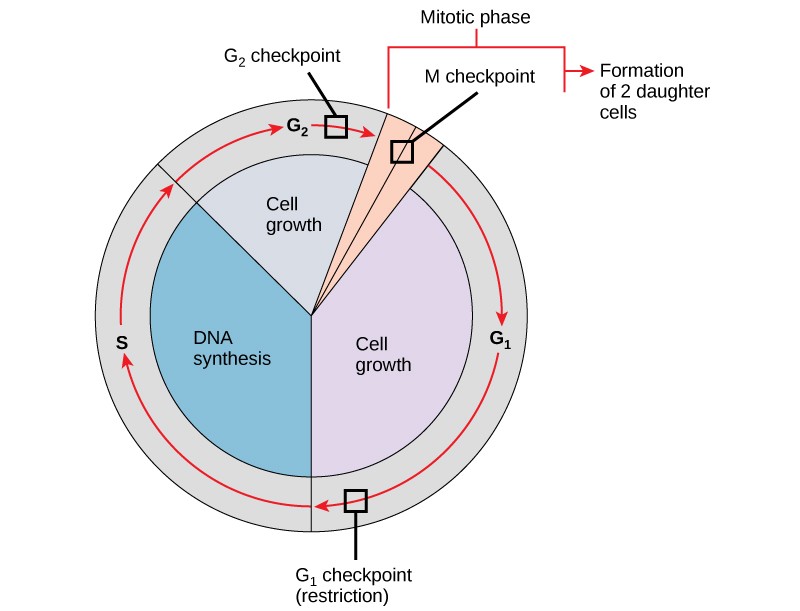
The G1 Checkpoint
The G1 checkpoint determines whether all conditions are favorable for cell division to proceed. The G1 checkpoint, also called the restriction point (in yeast), is a point at which the cell irreversibly commits to the cell division process. External influences, such as growth factors, play a large role in carrying the cell past the G1 checkpoint. In addition to adequate reserves and cell size, there is a check for genomic DNA damage at the G1 checkpoint. A cell that does not meet all the requirements will not be allowed to progress into the S phase. The cell can halt the cycle and attempt to remedy the problematic condition, or the cell can advance into G0 and await further signals when conditions improve.
The G2 Checkpoint
The G2 checkpoint bars entry into the mitotic phase if certain conditions are not met. As at the G1 checkpoint, cell size and protein reserves are assessed. However, the most important role of the G2 checkpoint is to ensure that all of the chromosomes have been replicated and that the replicated DNA is not damaged. If the checkpoint mechanisms detect problems with the DNA, the cell cycle is halted, and the cell attempts to either complete DNA replication or repair the damaged DNA.
The M Checkpoint
The M checkpoint occurs near the end of the metaphase stage of mitosis. The M checkpoint is also known as the spindle checkpoint, because it determines whether all of the sister chromatids are correctly attached to the spindle microtubules. Because the separation of the sister chromatids during anaphase is an irreversible step, the cycle will not proceed until the kinetochores of each pair of sister chromatids are firmly anchored to at least two spindle fibers arising from opposite poles of the cell.
13.3.3 Regulator Molecules of the Cell Cycle
In addition to the internally controlled checkpoints, there are two groups of intracellular molecules that regulate the cell cycle. These regulatory molecules either promote progress of the cell to the next phase (positive regulation) or halt the cycle (negative regulation). Regulator molecules may act individually, or they can influence the activity or production of other regulatory proteins. Therefore, the failure of a single regulator may have almost no effect on the cell cycle, especially if more than one mechanism controls the same event. Conversely, the effect of a deficient or non-functioning regulator can be wide-ranging and possibly fatal to the cell if multiple processes are affected.
Positive Regulation of the Cell Cycle
Two groups of proteins, called cyclins and cyclin-dependent kinases (Cdks), are responsible for the progress of the cell through the various checkpoints. The levels of the four cyclin proteins fluctuate throughout the cell cycle in a predictable pattern (Figure 13.11). Increases in the concentration of cyclin proteins are triggered by both external and internal signals. After the cell moves to the next stage of the cell cycle, the cyclins that were active in the previous stage are degraded.
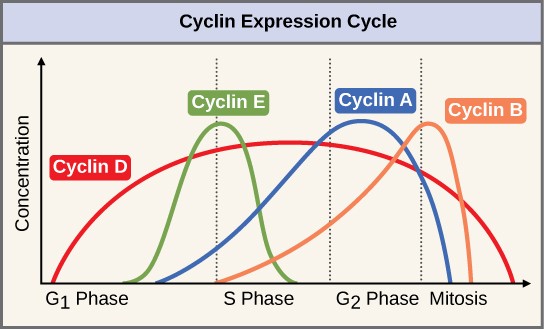
Cyclins regulate the cell cycle only when they are tightly bound to Cdks. To be fully active, the Cdk/cyclin complex must also be phosphorylated in specific locations. Like all kinases, Cdks are enzymes (kinases) that phosphorylate other proteins. Phosphorylation activates the protein by changing its shape. The proteins phosphorylated by Cdks are involved in advancing the cell to the next phase. (Figure 13.12). The levels of Cdk proteins are relatively stable throughout the cell cycle; however, the concentrations of cyclin fluctuate and determine when Cdk/cyclin complexes form. The different cyclins and Cdks bind at specific points in the cell cycle and thus regulate different checkpoints.
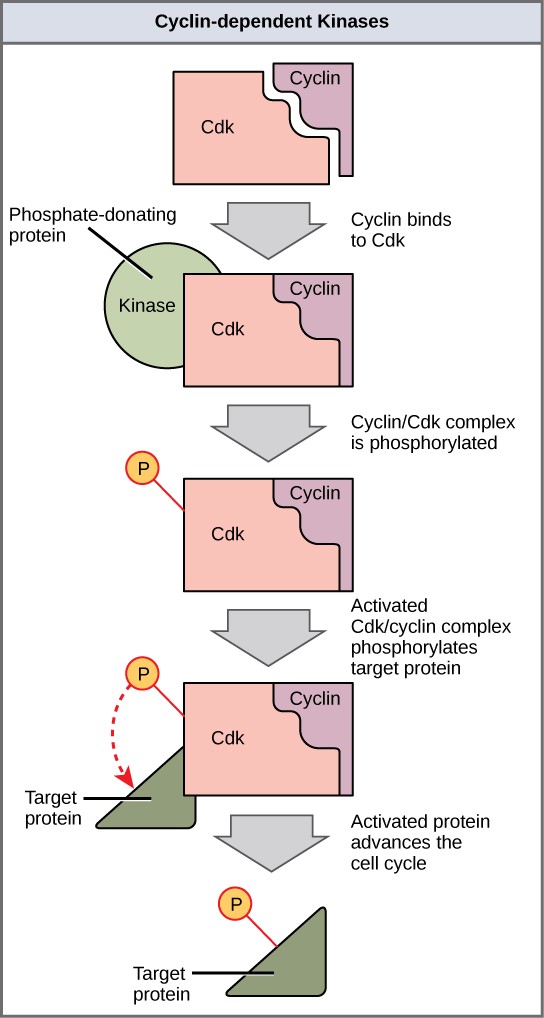
Since the cyclic fluctuations of cyclin levels are based on the timing of the cell cycle and not on specific events. Regulation of the cell cycle usually occurs by either the Cdk molecules alone or the Cdk/cyclin complexes. Without a specific concentration of fully activated cyclin/Cdk complexes, the cell cycle cannot proceed through the checkpoints.
Although the cyclins are the main regulatory molecules that determine the forward momentum of the cell cycle, there are several other mechanisms that fine-tune the progress of the cycle with negative, rather than positive, effects. These mechanisms essentially block the progression of the cell cycle until problematic conditions are resolved. Molecules that prevent the full activation of Cdks are called Cdk inhibitors. Many of these inhibitor molecules directly or indirectly monitor a particular cell cycle event. The block placed on Cdks by inhibitor molecules will not be removed until the specific event that the inhibitor monitors is completed.
Negative Regulation of the Cell Cycle
The second group of cell cycle regulatory molecules are negative regulators. Negative regulators halt the cell cycle. Remember that in positive regulation, active molecules cause the cycle to progress.
The best understood negative regulatory molecules are retinoblastoma protein (Rb), p53, and p21. Retinoblastoma proteins are a group of tumor-suppressor proteins common in many cells. The 53 and 21 designations refer to the functional molecular masses of the proteins (p) in kilodaltons. Much of what is known about cell cycle regulation comes from research conducted with cells that have lost regulatory control. All three of these regulatory proteins were discovered to be damaged or non-functional in cells that had begun to replicate uncontrollably (became cancerous). In each case, the main cause of the unchecked progress through the cell cycle was a faulty copy of the regulatory protein.
Rb, p53, and p21 act primarily at the G1 checkpoint. Regulator protein p53 is a multi-functional protein that has a major impact on the commitment of a cell to division because it acts when there is damaged DNA in cells that are undergoing the preparatory processes during G1. If damaged DNA is detected, p53 halts the cell cycle and recruits enzymes to repair the DNA. If the DNA cannot be repaired, p53 can trigger apoptosis, or cell suicide, to prevent the duplication of damaged chromosomes. As p53 levels rise, the production of p21 is triggered. Regulator protein p21 enforces the halt in the cycle dictated by p53 by binding to and inhibiting the activity of the Cdk/cyclin complexes. As a cell is exposed to more stress, higher levels of p53 and p21 will accumulate, making it less likely that the cell will move into the S phase.
Rb exerts its regulatory influence on other positive regulator proteins. Chiefly, Rb monitors cell size. In the active, dephosphorylated state, Rb binds to proteins called transcription factors, most commonly, E2F (Figure 13.13). Transcription factors “turn on” specific genes, allowing the production of proteins encoded by that gene. When Rb is bound to E2F, production of proteins necessary for the G1/S transition is blocked. As the cell increases in size, Rb is slowly phosphorylated until it becomes inactivated. Rb releases E2F, which can now turn on the gene that produces the transition protein, and this particular block is removed. For the cell to move past each of the checkpoints, all positive regulators must be “turned on,” and all negative regulators must be “turned off.”
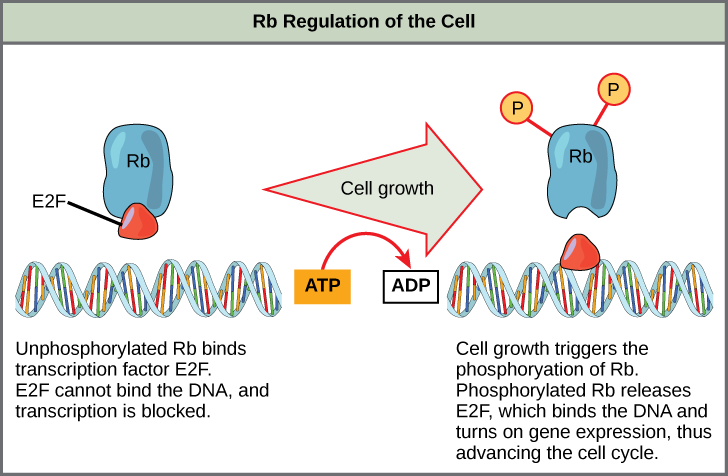
13.4 | Cancer and the Cell Cycle
Learning Objectives
By the end of this section, you will be able to:
- Describe how cancer is caused by uncontrolled cell growth.
- Understand how proto-oncogenes are normal cell genes that, when mutated, become oncogenes.
- Describe how tumor suppressors function.
- Explain how mutant tumor suppressors cause cancer.
Cancer comprises many different diseases caused by a common mechanism: uncontrolled cell growth. Despite the overlapping levels of cell cycle control, errors do occur. One of the critical processes monitored by the cell cycle checkpoint surveillance mechanism is the proper replication of DNA during the S phase. Even when all of the cell cycle controls are fully functional, a small percentage of replication errors (mutations) will be passed on to the daughter cells. When changes to a DNA nucleotide sequence occur within a coding portion of a gene and are not corrected, a gene mutation results.
All cancers start when a gene mutation gives rise to a faulty protein that plays a key role in cell reproduction. The change in the cell that results from the malformed protein may be minor: perhaps a slight delay in the binding of Cdk to cyclin or an Rb protein that detaches from its target DNA while still phosphorylated. Even minor mistakes may allow subsequent mistakes to occur more readily. Over and over, small uncorrected errors are passed from the parent cell to the daughter cells and amplified as each generation produces more non-functional proteins from uncorrected DNA damage. Eventually, the pace of the cell cycle speeds up as the effectiveness of the control and repair mechanisms decreases. Uncontrolled growth of the mutated cells outpaces the growth of normal cells in the area, and a tumor can result.
13.4.1 Proto-oncogenes
The genes that code for the positive cell cycle regulators are called proto-oncogenes. Proto-oncogenes are normal genes that, when mutated in certain ways, become oncogenes, genes that cause a cell to become cancerous. Consider what might happen to the cell cycle in a cell with a recently acquired oncogene. In most instances, the alteration of the DNA sequence will result in a less functional (or non-functional) protein. The result is detrimental to the cell and will likely prevent the cell from completing the cell cycle; however, the organism is not harmed because the mutation will not be carried forward. If a cell cannot reproduce, the mutation is not propagated and the damage is minimal. Occasionally, a gene mutation causes a change that increases the activity of a positive regulator. For example, a mutation that allows Cdk to be activated without being partnered with cyclin could push the cell cycle past a checkpoint before all of the required conditions are met. If the resulting daughter cells are too damaged to undergo further cell divisions, the mutation would not be propagated and no harm would come to the organism. If the atypical daughter cells are able to undergo further cell divisions, subsequent generations of cells will probably accumulate even more mutations, some possible in additional genes that regulate the cell cycle.
The Cdk gene in the above example is only one of many genes that are considered proto-oncogenes. In addition to the cell cycle regulatory proteins, any protein that influences the cycle can be altered in such a way as to override cell cycle checkpoints. An oncogene is any gene that, when altered, leads to an increase in the rate of cell cycle progression.
13.4.2 Tumor Suppressor Genes
Like proto-oncogenes, many of the negative cell cycle regulatory proteins were discovered in cells that had become cancerous. Tumor suppressor genes are segments of DNA that code for negative regulator proteins, the type of regulators that, when activated, can prevent the cell from undergoing uncontrolled division. The collective function of the best-understood tumor suppressor gene proteins, Rb, p53, and p21, is to put up a roadblock to cell cycle progression until certain events are completed. A cell that carries a mutated form of a negative regulator might not be able to halt the cell cycle if there is a problem. Tumor suppressors are similar to brakes in a vehicle: Malfunctioning brakes can contribute to a car crash.
Mutated p53 genes have been identified in more than one-half of all human tumor cells. This discovery is not surprising in light of the multiple roles that the p53 protein plays at the G1 checkpoint. A cell with a faulty p53 may fail to detect errors present in the genomic DNA (Figure 13.14). Even if a partially functional p53 does identify the mutations, it may no longer be able to signal the necessary DNA repair enzymes. Either way, damaged DNA will remain uncorrected. At this point, a functional p53 will deem the cell unsalvageable and trigger programmed cell death (apoptosis). The damaged version of p53 found in cancer cells cannot trigger apoptosis.
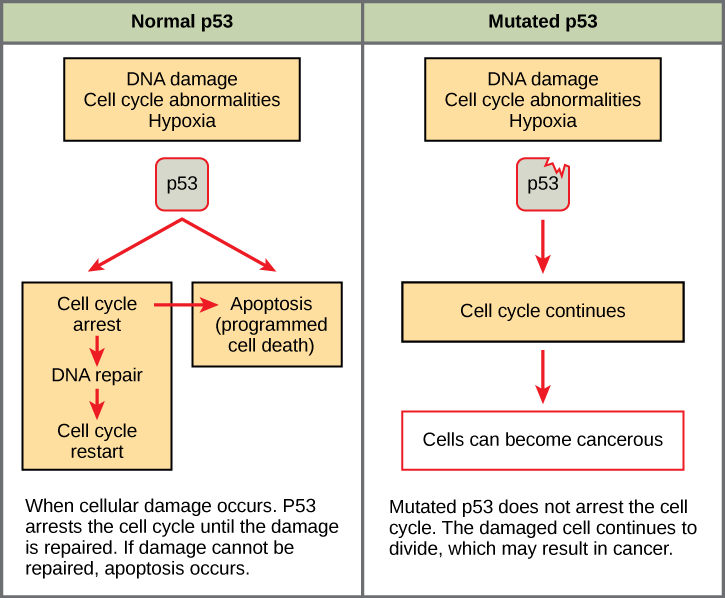
The loss of p53 function has other repercussions for the cell cycle. Mutated p53 might lose its ability to trigger p21 production. Without adequate levels of p21, there is no effective block on Cdk activation. Essentially, without a fully functional p53, the G1 checkpoint is severely compromised and the cell proceeds directly from G1 to S regardless of internal and external conditions. At the completion of this shortened cell cycle, two daughter cells are produced that have inherited the mutated p53 gene. Given the non-optimal conditions under which the parent cell reproduced, it is likely that the daughter cells will have acquired other mutations in addition to the faulty tumor suppressor gene. Cells such as these daughter cells quickly accumulate both oncogenes and non-functional tumor suppressor genes. Again, the result is tumor growth.
13.5 | Prokaryotic Cell Division
Learning Objectives
By the end of this section, you will be able to:
- Describe the process of binary fission in prokaryotes.
- Explain how FtsZ and tubulin proteins are examples of homology.
Prokaryotes propagate by binary fission. For unicellular organisms, cell division is the only method to produce new individuals. In both prokaryotic and eukaryotic cells, the outcome of cell reproduction is a pair of daughter cells that are genetically identical to the parent cell. In unicellular organisms, daughter cells are individuals. To achieve the outcome of cloned offspring, certain steps are essential. The genomic DNA must be replicated and then allocated into the daughter cells; the cytoplasmic contents must also be divided to give both new cells the machinery to sustain life. In bacterial cells, the genome consists of a single, circular DNA chromosome; therefore, the process of cell division is greatly simplified. Karyokinesis is unnecessary because there is no nucleus and thus no need to direct one copy of the multiple chromosomes into each daughter cell.
13.5.1 Binary Fission
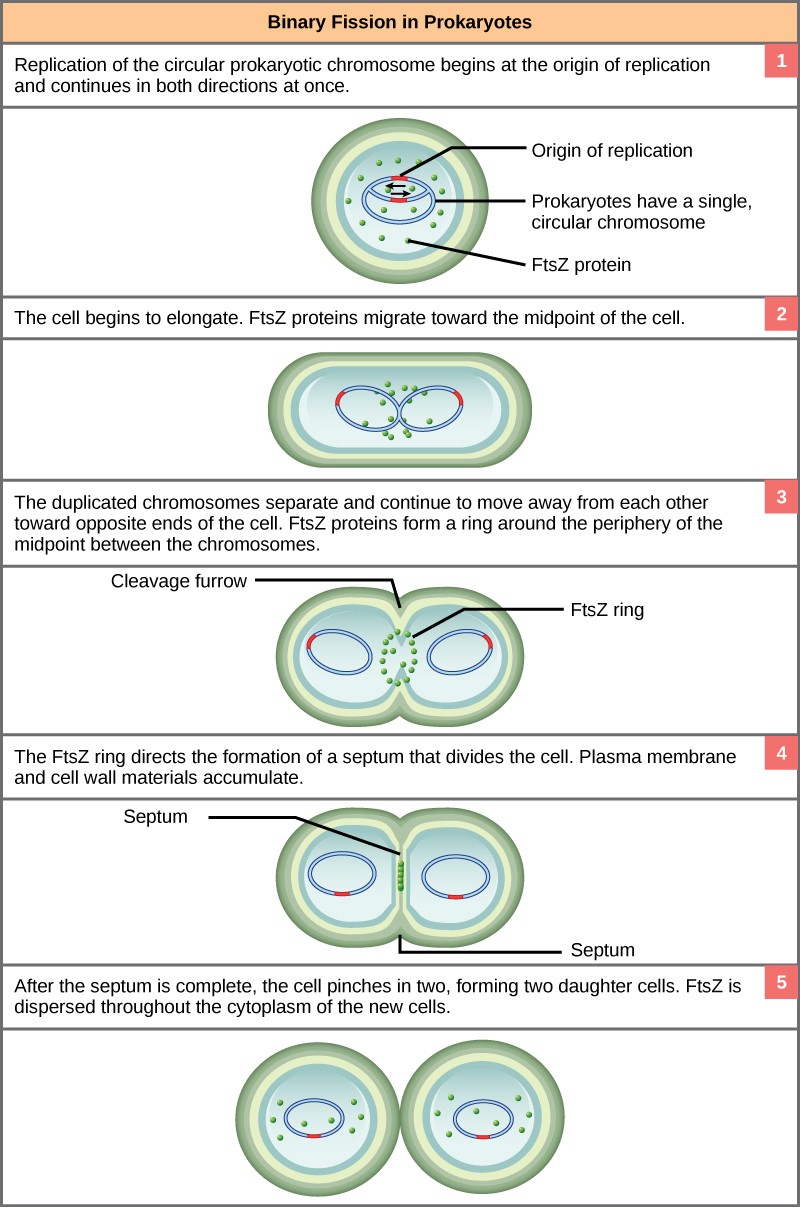
Due to the relative simplicity of the prokaryotes, the cell division process, called binary fission, is a less complicated and much more rapid process than cell division in eukaryotes. The single, circular DNA chromosome of bacteria is not enclosed in a nucleus, but instead occupies a specific location, the nucleoid, within the cell (Figure 13.2). Although the DNA of the nucleoid is associated with proteins that aid in packaging the molecule into a compact size, there are no histone proteins and thus no nucleosomes in prokaryotes. The packing proteins of bacteria are related to the cohesin and condensin proteins involved in the chromosome compaction of eukaryotes.
The bacterial chromosome is attached to the plasma membrane at about the midpoint of the cell. The starting point of replication, the origin, is close to the binding site of the chromosome to the plasma membrane (Figure 13.15). Replication of the DNA is bidirectional, moving away from the origin on both strands of the loop simultaneously. As the new double strands are formed, each origin point moves away from the plasma membrane attachment toward the opposite ends of the cell. As the cell elongates, the growing membrane aids in the transport of the chromosomes. After the chromosomes have cleared the midpoint of the elongated cell, cytoplasmic separation begins. The formation of a ring composed of repeating units of a protein called FtsZ directs the partition, cell wall, between the nucleoids. Formation of the FtsZ ring triggers the accumulation of other proteins that work together to recruit new membrane and cell wall materials to the site. A septum is formed between the nucleoids, extending gradually from the periphery toward the center of the cell. When the new cell walls are in place, the daughter cells separate.
Mitotic Spindle Apparatus
The precise timing and formation of the mitotic spindle is critical to the success of eukaryotic cell division. Prokaryotic cells, on the other hand, do not undergo karyokinesis and therefore have no need for a mitotic spindle. The FtsZ protein that plays such a vital role in prokaryotic cytokinesis is structurally and functionally very similar to tubulin, the building block of microtubules that make up the mitotic spindle fibers that are necessary for eukaryotes. FtsZ proteins can form filaments, rings, and other three-dimensional structures that resemble the way tubulin forms microtubules, centrioles, and various cytoskeletal components. In addition, both FtsZ and tubulin employ the same energy sourse, GTP (guanosine triphosphate), to rapidly assemble and disassemble complex structures.
FtsZ and tubulin are homologous structures derived from common evolutionary origins. In this example, FtsZ is the ancestor protein to tubulin (a modern protein). While both proteins are found in extant organisms, tubulin function has evolved and diversified tremendously since evolving from its FtsZ prokaryotic origin.

