2 Chapter 17. Regulation of Gene Expression
Chapter Outline
- 17.1 Overview of Regulation of Gene Expression
- 17.2 Prokaryotic Gene Regulation
- 17.3 Eukaryotic Gene Regulation
Introduction
Each somatic cell in the body generally contains the same DNA. (A few exceptions include red blood cells, which contain no DNA in their mature state, and some immune system cells that rearrange their DNA while producing antibodies.) In general, the genes that determine whether you have green eyes or brown hair, or how fast you metabolize food are the same in eye cells and liver cells, even though these organs function quite differently. If each cell has the same DNA, how is it that cells differ in their structure and function? Why do cells in the eye differ so dramatically from cells in the liver?
Although each cell in your body contains the same DNA sequences, each cell does not turn on, or express, the same set of genes. In fact, only a small subset of proteins are made by any one cell. In other words, in any given cell, not all genes encoded in the DNA are transcribed into mRNA or translated into protein. Cells in the eye make a certain subset of proteins, and liver cells make a different subset of proteins. In addition, at different times, liver cells may make different subsets of liver proteins. The expression of specific genes is a highly regulated process with many levels and stages of control. This complexity ensures expression of each protein in the proper cells at the proper time.
17.1 | Overview of Regulation of Gene Expression
Learning Objectives
By the end of this section, you will be able to:
- Discuss why every cell does not express all of its genes.
- Describe some major differences between prokaryotic and eukaryotic gene regulation.
For a cell to function properly, necessary proteins must be synthesized at the proper time. All cells control or regulate the synthesis of proteins from information encoded in their DNA. The process of “turning on” a gene to produce mRNA and protein is called gene expression. Whether in a simple unicellular organism or a complex multi-cellular organism, each cell controls when its genes are expressed, how much of the protein is made, and when it is time to stop making that protein because it is no longer needed.
The regulation of gene expression conserves energy and space. It is more energy efficient to turn on the genes only when they are required. In addition, only expressing a subset of genes in each cell saves space because DNA must be unwound from its tightly coiled structure to transcribe and translate the DNA. Cells would have to be enormous if every protein were expressed in every cell all the time. The control of gene expression is extremely complex. Malfunctions in this process are detrimental to the cell and can lead to the development of many diseases, including cancer.
17.1.1 Prokaryotic versus Eukaryotic Gene Expression
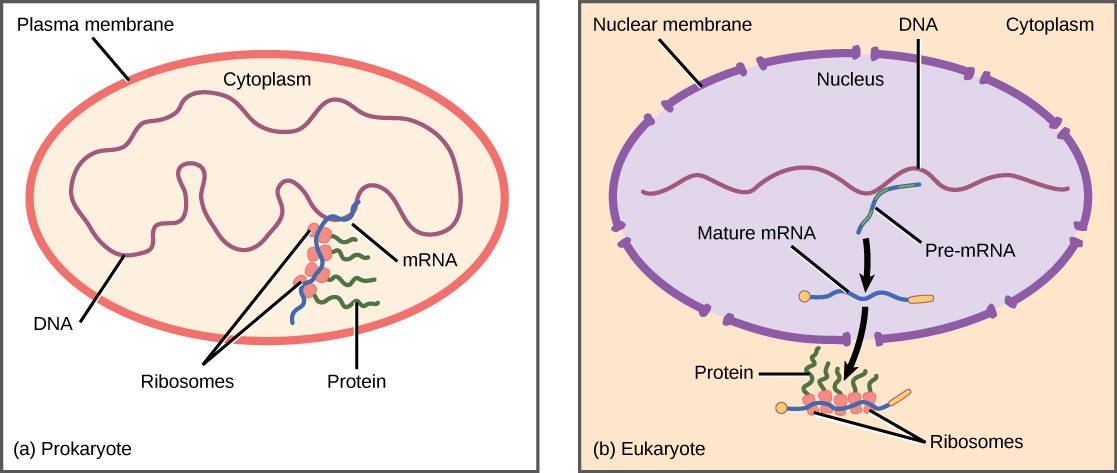
Since prokaryotic organisms are single-celled organisms that lack a cell nucleus, their DNA floats freely in the cell’s cytoplasm. When a particular protein is needed, the gene that codes for it is transcribed in mRNA, which is simultaneously translated into protein. When the protein is no longer needed, transcription stops. As a result, the primary method to control how much of each protein is expressed in a prokaryotic cell is the regulation of transcription.
Eukaryotic cells, in contrast, have intracellular organelles that add to their complexity. In eukaryotic cells, the DNA is contained inside the cell’s nucleus, where it is transcribed into mRNA. The newly synthesized mRNA is then modified and transported out of the nucleus into the cytoplasm, where ribosomes translate the mRNA into protein. The processes of transcription and translation are physically separated by the nuclear membrane; transcription occurs only within the nucleus, and translation occurs only in the cytoplasm. The regulation of gene expression in eukaryotes can occur at all stages of the process (Figure 17.2).
Some of the differences in the regulation of gene expression between prokaryotes and eukaryotes are summarized in Table 17.1.
Table 17.1 Differences in prokaryotic and eukaryotic gene regulation.
|
Prokaryotic organisms |
Eukaryotic organisms |
|
Lack nucleus |
Contain nucleus |
|
DNA is found in the cytoplasm |
DNA is in the nucleus |
|
Transcription and translation occur almost simultaneously |
Transcription occurs in the nucleus prior to translation, which occurs in the cytoplasm. |
|
Gene expression is regulated primarily at the transcriptional level |
Gene expression is regulated at many levels: epigenetic, transcriptional, nuclear shuttling, post-transcriptional, translational, and post-translational |
17.2 | Prokaryotic Gene Regulation
Learning Objectives
By the end of this section, you will be able to:
- Describe the steps involved in prokaryotic gene regulation.
- Explain the roles of activators, inducers, and repressors in gene regulation
The DNA of prokaryotes is organized into a circular chromosome that resides in the cell’s cytoplasm. Proteins that are needed for a specific function, or that are involved in the same biochemical pathway, are often encoded together in blocks called operons. For example, all five of the genes needed to make the amino acid tryptophan in the bacterium E. coli are located next to each other in the trp operon. The genes in an operon are transcribed into a single mRNA molecule. This allows the genes to be controlled as a unit: either all are expressed, or none is expressed. Each operon needs only one regulatory region, including a promoter, where RNA polymerase binds, and an operator, where other regulatory proteins bind.
In prokaryotic cells, there are three types of regulatory molecules that can affect the expression of operons. Activators are proteins that increase the transcription of a gene. Repressors are proteins that suppress transcription of a gene. Finally, inducers are molecules that bind to repressors and inactivate them. Below are two examples of how these molecules regulate different operons.
17.2.1 The trp Operon: A Repressor Operon
Like all cells, bacteria need amino acids to survive. Tryptophan is one amino acid that the bacterium E. coli can either ingest from the environment or synthesize. When E. coli needs to synthesize tryptophan, it must express a set of five proteins that are encoded by five genes. These five genes are located next to each other in the tryptophan (trp) operon (Figure 17.3).
When tryptophan is present in the environment, E. coli does not need to synthesize it, and the trp operon is switched off. However, when tryptophan availability is low, the trp operon is turned on so that the genes are transcribed, the proteins are made, and tryptophan can be synthesized.
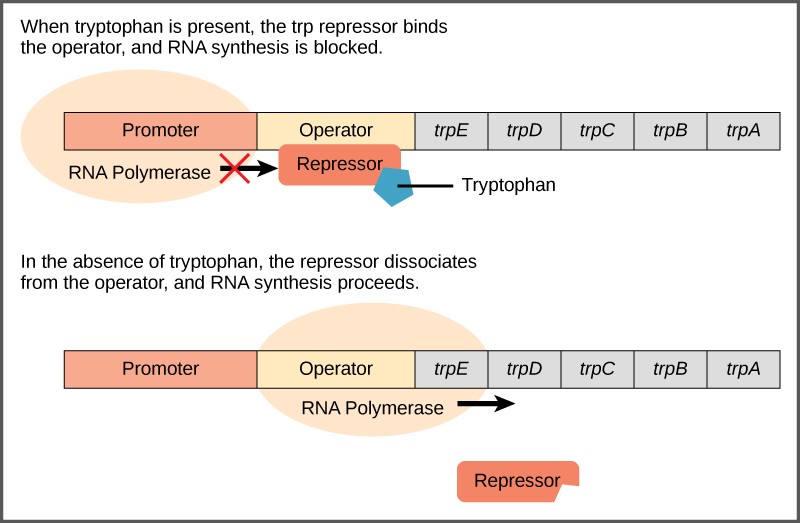
A DNA sequence called the operator is located between the promoter and the first trp gene. The operator contains the DNA code to which the repressor protein can bind. The repressor protein is regulated by levels of tryptophan in the cell.
When tryptophan is present in the cell, two tryptophan molecules bind to the trp repressor. This causes the repressor to change shape and bind to the trp operator. Binding of the tryptophan–repressor complex at the operator physically blocks the RNA polymerase from binding, and transcribing the downstream genes. Thus, when the cell has enough tryptophan, it is preventing from making more.
When tryptophan is not present in the cell, the repressor has no tryptophan to bind to it. The repressor is not activated and it does not bind to the operator. Therefore, RNA polymerase can transcribe the operon and make the enzymes to synthesize tryptophan. Thus, when the cell does not have enough tryptophan, it synthesizes it.
17.2.2 The lac Operon: An Inducer Operon
The lac operon in E. coli has more complex regulation, involving both a repressor and an activator. E. coli uses glucose for food, but is able to use other sugars, such as lactose, when glucose concentrations are low. Three proteins are needed to break down lactose; they are encoded by the three genes of the lac operon.
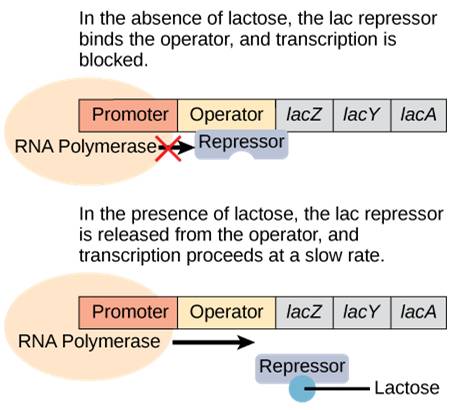
When lactose is not present, the proteins to digest lactose are not needed. Therefore, a repressor binds to the operator and prevents RNA polymerase from transcribing the operon.
When lactose is present, lactose binds to the repressor and removes it from the operator. RNA polymerase is now free to transcribe the genes necessary to digest lactose (Figure 17.4)
However, the story is more complex than this. Since E. coli prefers to use glucose for food, the lac operon is only expressed at low levels even when the repressor is removed. But what happens when ONLY lactose is present? Now the bacterium needs to ramp up production of the lactose-digesting proteins. It does so by using an activator protein called catabolite activator protein (CAP).
When glucose levels drop, cyclic AMP (cAMP) begins to accumulate in the cell. cAMP binds to CAP and the complex binds to the lac operon promoter (Figure 17.5). This increases the binding ability of RNA polymerase to the promoter and ramps up transcription of the genes.
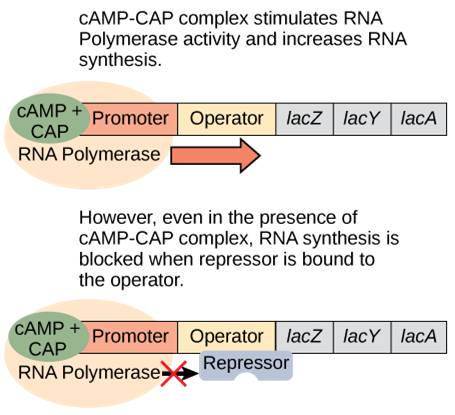
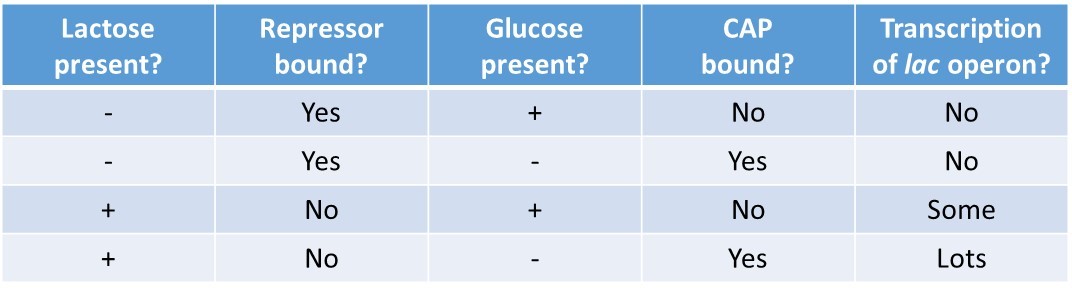
In summary, for the lac operon to be fully activated, two conditions must be met. First, the level of glucose must be very low or non-existent. Second, lactose must be present. Only when glucose is absent and lactose is present will the lac operon be transcribed maximally. This makes sense for the cell, because it would be energetically wasteful to create the proteins to process lactose if glucose was plentiful or lactose was not available (Table 17.2).
Table 17.2 Summary of signals that induce or repress transcription of the lac operon.
15.3 Eukaryotic Gene Regulation
Learning Objectives
By the end of this section, you will be able to:
- Explain the process of epigenetic gene regulation in eukaryotic cells.
- Explain the process of transcriptional gene regulation in eukaryotic cells.
- Explain the process of post-transcriptional gene regulation in eukaryotic cells.
- Explain the process of translational gene regulation in eukaryotic cells.
- Explain the process of post-transcriptional gene regulation in eukaryotic cells.
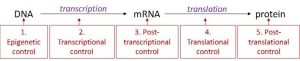
In eukaryotes, control of gene expression is more complex and can happen at many different levels. Eukaryotic genes are not organized into operons, so each gene must be regulated independently. In addition, eukaryotic cells have many more genes than prokaryotic cells. Regulation of gene expression can happen at any of the stages as DNA is transcribed into mRNA and mRNA is translated into protein. For convenience, regulation is divided into five levels: epigenetic, transcriptional, post-transcriptional, translational, and post-translational (Figure 17.6).
17.3.1 Epigenetic Control fo Gene Expression
The first level of control of gene expression is epigenetic (“around genetics”) regulation. Epigenetics is a relatively new, but growing, field of biology.
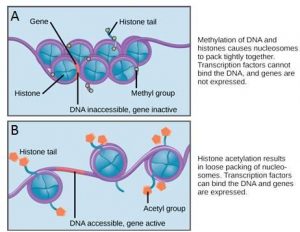
Epigenetic control involves changes to genes that do not alter the nucleotide sequence of the DNA and are not permanent. Instead, these changes alter the chromosomal structure so that genes can be turned on or off. This level of control occurs through heritable chemical modifications of the DNA and/or chromosomal proteins.
One example of chemical modifications of DNA is the addition of methyl groups to the DNA, in a process called methylation, In general, methylation suppresses transcription. Interestingly, methylation patterns can be passed on as cells divide. Thus, parents may be able to pass on the tendency of a gene to be expressed in their offspring. Other heritable chemical modifications of DNA may also occur.
Modification of Histone Proteins is an Example of Epigenetic Control
The best-studied example of epigenetic regulation is modification of histone proteins. Histones are chromosomal proteins that tightly wind DNA so that it fits into the nucleus of a cell. The human genome, for example, consists of over three billion nucleotide pairs. An average chromosome contains 130 million nucleotide pairs, and each body cell contains 46 chromosomes. If stretched out linearly, an average human chromosome would be over four centimeters long. In order to fit all of this DNA into the nucleus of a microscopic cell, the DNA must be tightly wound around proteins. It is also organized so that specific segments can be accessed as needed by a specific cell type (Figure 17.7).
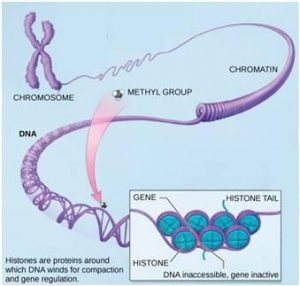
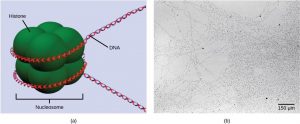
The first level of organization, or packing, is the winding of DNA strands around histone proteins. Histones package and order DNA into structural units called nucleosome complexes, which can control the access of proteins to the DNA regions (Figure 17.8a). Under the electron microscope, this winding of DNA around histone proteins to form nucleosomes looks like small beads on a string (Figure 17.8b). These beads (histone proteins) can move along the string (DNA) and change the structure of the molecule.
If a gene is to be transcribed, the nucleosomes surrounding that region of DNA can slide down the DNA to open that specific chromosomal region and allow access for RNA polymerase and other proteins, called transcription factors, to bind to the promoter region and initiate transcription. If a gene is to remain turned off, or silenced, the histone proteins and DNA have different modifications that signal a closed chromosomal configuration. In this closed configuration, the RNA polymerase and transcription factors do not have access to the DNA and transcription cannot occur (Figure 17.9).
How the histone proteins move is dependent on signals found on the histone proteins. These signals are “tags” – in the form of phosphate, methyl, or acetyl groups – that open or close a chromosomal region (Figure 17.9). These tags are not permanent, but may be added or removed as needed. Since DNA negatively charged, changes in the charge of the histone will change how tightly wound the DNA molecule will be. When unmodified, the histone proteins have a large positive charge; by adding chemical modifications like acetyl groups, the charge becomes less positive.
17.3.2 Transcriptional Control of Gene Expression
Transcriptional regulation is control of whether or not an mRNA is transcribed from a gene in a particular cell. Like prokaryotic cells, the transcription of genes in eukaryotes requires an RNA polymerase to bind to a promoter to initiate transcription. In eukaryotes, RNA polymerase requires other proteins, or transcription factors, to facilitate transcription initiation. Transcription factors are proteins that bind to the promoter sequence and other regulatory sequences to control the transcription of the target gene. RNA polymerase by itself cannot initiate transcription in eukaryotic cells. Transcription factors must bind to the promoter region first and recruit RNA polymerase to the site for transcription to begin.
The Promoter and Transcription Factors
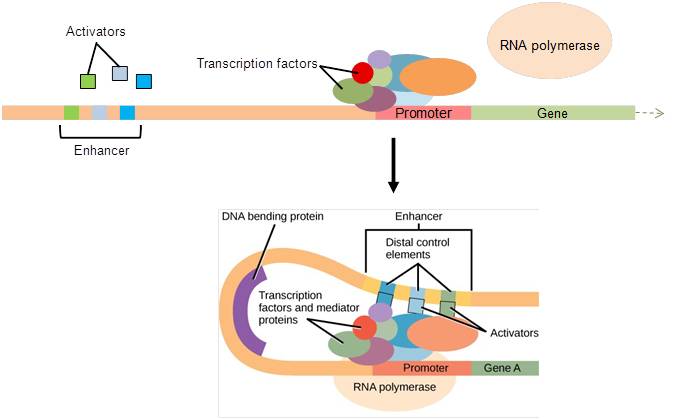
In eukaryotic genes, the promoter region is immediately upstream of the coding sequence. This region can range from a few to hundreds of nucleotides long. The length of the promoter is gene-specific and can differ dramatically between genes. The longer the promoter, the more available space for proteins to bind. Consequently, the level of control of gene expression can differ quite dramatically between genes. The purpose of the promoter is to bind transcription factors that control the initiation of transcription (Figure 17.10, top).
Within the promoter region, just upstream of the transcriptional start site, resides the TATA box. This box is simply a repeat of thymine and adenine dinucleotides (literally, TATA repeats). Transcription factors bind to the TATA box, assembling an initiation complex. Once this complex is assembled, RNA polymerase binds to its upstream sequence and becomes phosphorylated. This releases part of the protein from the DNA, activates the transcription initiation complex, and places RNA polymerase in the correct orientation to begin transcription (Figure 17.10, top).
Enhancers and Repressors
In some eukaryotic genes, there are regions that help increase transcription. These regions, called enhancers, are not necessarily close to the genes; they can be located thousands of nucleotides away. They can be found upstream, within the coding region, or downstream of a gene. Enhancers are binding sites for activators. When an enhancer is far away from a gene, the DNA folds such that the enhancer is brought into proximity with the promoter, allowing interaction between the activators and the transcription initiation complex (Figure 17.10, bottom).
Like prokaryotic cells, eukaryotic cells also have mechanisms to prevent transcription. Transcriptional repressors can bind to promoter or enhancer regions and block transcription. Both activators and repressors respond to external stimuli to determine which genes need to be expressed.
17.3.3 Post-transcriptional Control of Gene Expression
Post-transcriptional regulation occurs after the mRNA is transcribed but before translation begins. This regulation can occur at the level of mRNA processing, transport from the nucleus to the cytoplasm, or binding to ribosomes.
Alternative RNA splicing
Recall from chapter 5 that in eukaryotic cells the RNA primary transcript often contains introns, which are removed prior to translation.
Alternative RNA splicing is a mechanism that allows different combinations of introns, and sometimes exons, to be removed from the primary transcript (Figure 17.11). This allows different protein products to be produced from one gene. Alternative splicing can act as a mechanism of gene regulation. Differential splicing is used to produce different protein products in different cells or at different times within the same cell. Alternative splicing is now understood to be a common mechanism of gene regulation in eukaryotes; up to 70 percent of genes in humans are expressed as multiple proteins through alternative splicing.
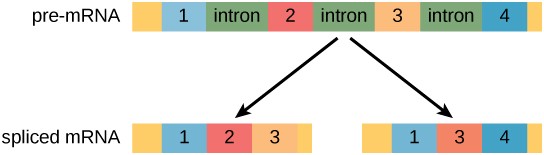
Evolution of Alternative Splicing
How could alternative splicing evolve? Introns have a beginning and ending recognition sequence; it is easy to imagine the failure of the splicing mechanism to identify the end of an intron and instead find the end of the next intron, thus removing two introns and the intervening exon. In fact, there are mechanisms in place to prevent such intron skipping, but mutations are likely to lead to their failure. Such “mistakes” would more than likely produce a nonfunctional protein. Indeed, the cause of many genetic diseases is alternative splicing rather than mutations in a sequence. However, alternative splicing would create a protein variant without the loss of the original protein, opening up possibilities for adaptation of the new variant to new functions. Gene duplication has played an important role in the evolution of new functions in a similar way by providing genes that may evolve without eliminating the original, functional protein.
Control of RNA Stability
Another type of post-transcriptional control involves the stability of the mRNA in the cytoplasm. The longer an mRNA exists in the cytoplasm, the more time it has to be translated, and the more protein is made. Many factors contribute to mRNA stability, including the length of its poly-A tail.
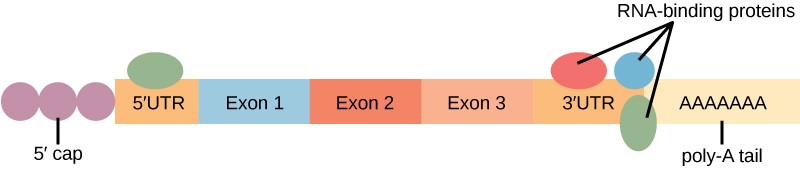
Proteins, called RNA-binding proteins (RBPs) can bind to the regions of the RNA just upstream or downstream of the protein-coding region. These regions in the RNA that are not translated into protein are called the untranslated regions, or UTRs. The region just before the protein-coding region is called the 5′ UTR, whereas the region after the coding region is called the 3′ UTR (Figure 17.12). The binding of RBPs to these regions can increase or decrease the stability of an RNA molecule, depending on the specific RBP that binds.
microRNAs, or miRNAs, can also bind to the RNA molecule. miRNAs are short (21–24 nucleotides) RNA molecules that are made in the nucleus as longer pre-miRNAs and then chopped into mature miRNAs by a protein called dicer. miRNAs bind to mRNA along with a ribonucleoprotein complex called the RNA-induced silencing complex (RISC). The RISC-miRNA complex rapidly degrades the target mRNA.
17.3.4 Translational Control of Gene Expression
After an mRNA has been transported to the cytoplasm, it is translated into proteins. Control of this process is largely dependent on the mRNA molecule. As previously discussed, the stability of the mRNA will have a large impact on its translation into a protein. Translation can also be regulated at the level of binding of the mRNA to the ribosome. Once the mRNA bound to the ribosome, the speed and level of translation can still be controlled. An example of translational control occurs in proteins that are destined to end up in an organelle called the endoplasmic reticulum (ER). The first few amino acids of these proteins are a tag called a signal sequence. As soon as these amino acids are translated, a signal recognition particle (SRP) binds to the signal sequence and stops translation while the mRNA-ribosome complex is shuttled to the ER. Once they arrive, the SRP is removed and translation resumes.
17.3.5 Post-translational Control of Gene Expression
The final level of control of gene expression in eukaryotes is post-translational regulation. This type of control involves modifying the protein after it is made, in such as way as to affect its activity. One example of post-translational regulation is enzyme inhibition. When an enzyme is no longer needed, it is inhibited by a competitive or allosteric inhibitor, which prevents it from binding to its substrate. The inhibition is reversible, so that the enzyme can be reactivated later. This is more efficient than degrading the enzyme when it is not needed and then making more when it is needed again.
The activity and/or stability of proteins can also be regulated by adding functional groups, such as methyl, phosphate, or acetyl groups. Sometimes these modifications can regulate where a protein is found in the cell—for example, in the nucleus, the cytoplasm, or attached to the plasma membrane.
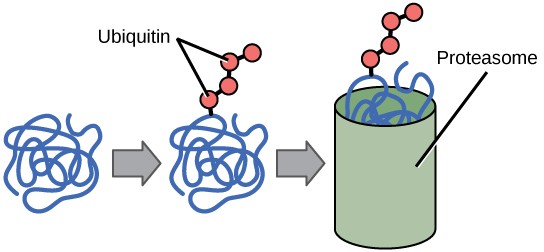
The addition of an ubiquitin group to a protein marks that protein for degradation. Ubiquitin acts like a flag indicating that the protein’s lifespan is complete. Tagged proteins are moved to a proteasome, an organelle that degrades proteins (Figure 17.13). One way to control gene expression, therefore, is to alter the longevity of the protein.